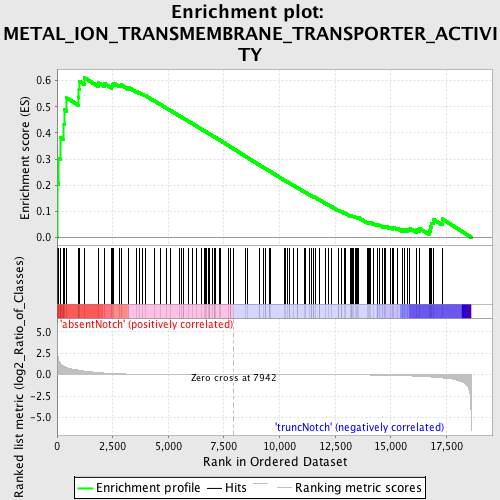
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | METAL_ION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY |
| Enrichment Score (ES) | 0.6115274 |
| Normalized Enrichment Score (NES) | 1.5556986 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.28649658 |
| FWER p-Value | 0.962 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2A3 | 130440 2190451 | 4 | 3.349 | 0.2072 | Yes | ||
| 2 | ITPR1 | 3450519 | 79 | 1.590 | 0.3017 | Yes | ||
| 3 | ATP2A2 | 1090075 3990279 | 134 | 1.356 | 0.3827 | Yes | ||
| 4 | SLC40A1 | 2370286 | 307 | 0.951 | 0.4323 | Yes | ||
| 5 | SLC31A1 | 4590161 6940154 | 323 | 0.929 | 0.4890 | Yes | ||
| 6 | SLC30A5 | 2630288 2970403 | 434 | 0.817 | 0.5337 | Yes | ||
| 7 | CACNG4 | 6980112 | 957 | 0.514 | 0.5373 | Yes | ||
| 8 | KCNH2 | 1170451 | 982 | 0.506 | 0.5673 | Yes | ||
| 9 | CACNA1H | 1230279 | 1011 | 0.498 | 0.5967 | Yes | ||
| 10 | SCN5A | 4070017 | 1218 | 0.420 | 0.6115 | Yes | ||
| 11 | CACNB3 | 2480035 4540184 | 1873 | 0.239 | 0.5910 | No | ||
| 12 | NOLA1 | 730093 | 2119 | 0.188 | 0.5894 | No | ||
| 13 | CACNB2 | 1500095 7330707 | 2462 | 0.132 | 0.5791 | No | ||
| 14 | KCNN3 | 6520600 6420138 | 2488 | 0.129 | 0.5857 | No | ||
| 15 | KCNN2 | 990025 3520524 | 2550 | 0.122 | 0.5900 | No | ||
| 16 | SLC31A2 | 1450577 | 2800 | 0.097 | 0.5826 | No | ||
| 17 | P2RX7 | 6020048 6380053 6510121 | 2880 | 0.091 | 0.5839 | No | ||
| 18 | ITPR2 | 5360128 | 3200 | 0.067 | 0.5708 | No | ||
| 19 | SLC8A1 | 3440717 | 3221 | 0.065 | 0.5737 | No | ||
| 20 | P2RX1 | 2940021 3290338 | 3557 | 0.046 | 0.5585 | No | ||
| 21 | KCNQ2 | 130204 290291 5860132 | 3684 | 0.040 | 0.5542 | No | ||
| 22 | SLC13A4 | 2940541 | 3836 | 0.034 | 0.5481 | No | ||
| 23 | SLC30A4 | 1850372 | 3953 | 0.030 | 0.5437 | No | ||
| 24 | RYR1 | 2510114 3130408 3710086 | 4355 | 0.020 | 0.5233 | No | ||
| 25 | KCNA2 | 5080403 | 4634 | 0.016 | 0.5092 | No | ||
| 26 | KCNC1 | 770110 | 4931 | 0.012 | 0.4940 | No | ||
| 27 | CHRNA6 | 5340092 | 5117 | 0.011 | 0.4846 | No | ||
| 28 | KCNIP2 | 60088 1780324 | 5497 | 0.008 | 0.4647 | No | ||
| 29 | KCNA6 | 450291 1400086 1660291 2470576 | 5572 | 0.008 | 0.4611 | No | ||
| 30 | CACNA1B | 1740010 5360593 | 5689 | 0.007 | 0.4553 | No | ||
| 31 | P2RX3 | 4230397 | 5698 | 0.007 | 0.4554 | No | ||
| 32 | PKD2 | 4670390 | 5922 | 0.006 | 0.4437 | No | ||
| 33 | ATP2B3 | 2260022 3780059 | 6094 | 0.006 | 0.4348 | No | ||
| 34 | CHRND | 840403 2260670 | 6264 | 0.005 | 0.4259 | No | ||
| 35 | KCNJ12 | 3870364 | 6500 | 0.004 | 0.4135 | No | ||
| 36 | KCNMB2 | 3780128 4760136 | 6603 | 0.004 | 0.4082 | No | ||
| 37 | KCNH1 | 4070619 6660181 | 6659 | 0.004 | 0.4055 | No | ||
| 38 | KCNJ10 | 2260577 | 6733 | 0.003 | 0.4017 | No | ||
| 39 | ACCN1 | 2060139 2190541 | 6792 | 0.003 | 0.3988 | No | ||
| 40 | CACNA1A | 6220164 | 6844 | 0.003 | 0.3962 | No | ||
| 41 | CUL5 | 450142 | 6869 | 0.003 | 0.3951 | No | ||
| 42 | KCNA5 | 5270750 | 6996 | 0.002 | 0.3884 | No | ||
| 43 | CHRNA1 | 1170025 3610364 5220292 | 7069 | 0.002 | 0.3847 | No | ||
| 44 | SLC39A2 | 1410338 | 7092 | 0.002 | 0.3836 | No | ||
| 45 | HTR3B | 430438 520301 | 7106 | 0.002 | 0.3830 | No | ||
| 46 | KCNS3 | 4150039 | 7305 | 0.002 | 0.3724 | No | ||
| 47 | KCNJ6 | 580465 5570246 | 7364 | 0.001 | 0.3694 | No | ||
| 48 | TRPC4 | 6040022 | 7707 | 0.001 | 0.3509 | No | ||
| 49 | KCNC4 | 110053 4760600 | 7778 | 0.000 | 0.3472 | No | ||
| 50 | KCNJ4 | 2350315 | 7780 | 0.000 | 0.3471 | No | ||
| 51 | CHRNA4 | 730075 2680091 | 7913 | 0.000 | 0.3400 | No | ||
| 52 | RYR2 | 2760671 | 8447 | -0.001 | 0.3113 | No | ||
| 53 | MS4A2 | 2940594 | 8571 | -0.002 | 0.3047 | No | ||
| 54 | SLC9A7 | 5360301 | 9074 | -0.003 | 0.2777 | No | ||
| 55 | RYR3 | 1660156 | 9110 | -0.003 | 0.2760 | No | ||
| 56 | SCN11A | 2480372 | 9286 | -0.003 | 0.2668 | No | ||
| 57 | KCNJ5 | 1570053 | 9372 | -0.004 | 0.2624 | No | ||
| 58 | KCNK3 | 2690372 | 9374 | -0.004 | 0.2626 | No | ||
| 59 | KCND3 | 670041 | 9549 | -0.004 | 0.2534 | No | ||
| 60 | KCNJ15 | 70176 6900750 | 9560 | -0.004 | 0.2531 | No | ||
| 61 | SCN4A | 5340687 | 9580 | -0.004 | 0.2524 | No | ||
| 62 | TRPC3 | 840064 | 10226 | -0.006 | 0.2179 | No | ||
| 63 | KCNJ3 | 50184 | 10266 | -0.006 | 0.2162 | No | ||
| 64 | KCNK1 | 1050068 | 10376 | -0.007 | 0.2107 | No | ||
| 65 | SCN9A | 5270575 | 10438 | -0.007 | 0.2078 | No | ||
| 66 | KCNMB1 | 4760139 | 10439 | -0.007 | 0.2082 | No | ||
| 67 | SCN7A | 2510538 | 10607 | -0.008 | 0.1997 | No | ||
| 68 | CACNB4 | 2030133 | 10785 | -0.008 | 0.1906 | No | ||
| 69 | KCNA4 | 2320706 | 10791 | -0.008 | 0.1908 | No | ||
| 70 | KCNK4 | 3170411 7040064 | 11115 | -0.009 | 0.1740 | No | ||
| 71 | SCNN1G | 520600 | 11149 | -0.009 | 0.1728 | No | ||
| 72 | SLC30A3 | 1850451 | 11349 | -0.010 | 0.1626 | No | ||
| 73 | CACNB1 | 2940427 3710487 | 11450 | -0.011 | 0.1579 | No | ||
| 74 | CHRNB2 | 580204 3120739 | 11453 | -0.011 | 0.1585 | No | ||
| 75 | ABCB11 | 7040170 | 11534 | -0.011 | 0.1549 | No | ||
| 76 | TRPV6 | 4010717 | 11544 | -0.011 | 0.1551 | No | ||
| 77 | KCNJ1 | 610707 870181 | 11594 | -0.012 | 0.1531 | No | ||
| 78 | CACNG5 | 1410746 2030673 | 11785 | -0.013 | 0.1436 | No | ||
| 79 | CHRNA3 | 6760100 | 11795 | -0.013 | 0.1439 | No | ||
| 80 | CACNA1S | 4730465 | 12070 | -0.014 | 0.1300 | No | ||
| 81 | CACNA1E | 840253 | 12219 | -0.015 | 0.1230 | No | ||
| 82 | HTR3A | 3310180 | 12352 | -0.017 | 0.1169 | No | ||
| 83 | KCNA1 | 2030594 | 12633 | -0.019 | 0.1029 | No | ||
| 84 | CHRNB3 | 430441 870725 5720204 | 12657 | -0.019 | 0.1029 | No | ||
| 85 | KCNE2 | 1780577 | 12763 | -0.020 | 0.0984 | No | ||
| 86 | TRPC1 | 3120189 4210537 | 12766 | -0.020 | 0.0996 | No | ||
| 87 | CACNA1C | 1660128 3780300 3870092 6290403 6900301 | 12898 | -0.022 | 0.0938 | No | ||
| 88 | CLDN16 | 1850286 | 12970 | -0.023 | 0.0914 | No | ||
| 89 | CACNA1D | 1740315 4730731 | 13198 | -0.025 | 0.0807 | No | ||
| 90 | KCNE1 | 1230520 1690372 4120050 | 13214 | -0.025 | 0.0814 | No | ||
| 91 | ABCC8 | 2810671 3120671 | 13225 | -0.025 | 0.0825 | No | ||
| 92 | SCNN1B | 5390377 | 13293 | -0.026 | 0.0805 | No | ||
| 93 | CHRNA7 | 2970446 | 13311 | -0.027 | 0.0812 | No | ||
| 94 | CACNA1G | 3060161 4810706 5550722 | 13403 | -0.028 | 0.0780 | No | ||
| 95 | KCNK7 | 4230576 | 13448 | -0.029 | 0.0774 | No | ||
| 96 | CHRNA2 | 5050315 | 13523 | -0.030 | 0.0753 | No | ||
| 97 | KCNQ3 | 1240037 | 13565 | -0.031 | 0.0750 | No | ||
| 98 | KCNS1 | 2570195 | 13953 | -0.038 | 0.0564 | No | ||
| 99 | TRPM2 | 460593 | 13992 | -0.039 | 0.0568 | No | ||
| 100 | TRPC5 | 3390270 | 14050 | -0.040 | 0.0562 | No | ||
| 101 | KCNC3 | 3440026 4670288 | 14092 | -0.041 | 0.0565 | No | ||
| 102 | SCN1B | 1240022 2120048 | 14210 | -0.044 | 0.0530 | No | ||
| 103 | CHRNE | 3190170 | 14380 | -0.049 | 0.0469 | No | ||
| 104 | ATP2C1 | 2630446 6520253 | 14382 | -0.049 | 0.0499 | No | ||
| 105 | CACNA2D1 | 5550593 | 14490 | -0.052 | 0.0473 | No | ||
| 106 | TRPM1 | 50746 2060301 5130100 5720129 | 14636 | -0.057 | 0.0430 | No | ||
| 107 | CACNA1F | 2360605 3390450 | 14736 | -0.060 | 0.0413 | No | ||
| 108 | CHRNB4 | 6370110 | 14772 | -0.062 | 0.0433 | No | ||
| 109 | KCNJ14 | 870441 4060113 6110372 | 14971 | -0.070 | 0.0369 | No | ||
| 110 | KCNA3 | 510059 | 15088 | -0.076 | 0.0354 | No | ||
| 111 | ACCN4 | 6380551 | 15101 | -0.077 | 0.0395 | No | ||
| 112 | KCNK5 | 4590504 6860687 | 15280 | -0.087 | 0.0353 | No | ||
| 113 | CACNG1 | 3060711 | 15503 | -0.103 | 0.0296 | No | ||
| 114 | ATP2A1 | 110309 | 15626 | -0.115 | 0.0301 | No | ||
| 115 | KCNH3 | 3130100 | 15764 | -0.129 | 0.0307 | No | ||
| 116 | KCNQ1 | 1580050 6860441 | 15860 | -0.139 | 0.0342 | No | ||
| 117 | SLC11A2 | 3140603 | 16164 | -0.180 | 0.0290 | No | ||
| 118 | P2RX4 | 2060497 6650162 | 16280 | -0.196 | 0.0349 | No | ||
| 119 | CCS | 3450524 | 16718 | -0.258 | 0.0272 | No | ||
| 120 | ITPR3 | 4010632 | 16780 | -0.270 | 0.0407 | No | ||
| 121 | FXN | 4070500 | 16840 | -0.279 | 0.0548 | No | ||
| 122 | KCNMB4 | 3290347 | 16905 | -0.292 | 0.0694 | No | ||
| 123 | KCNN4 | 2680129 | 17310 | -0.373 | 0.0706 | No |

