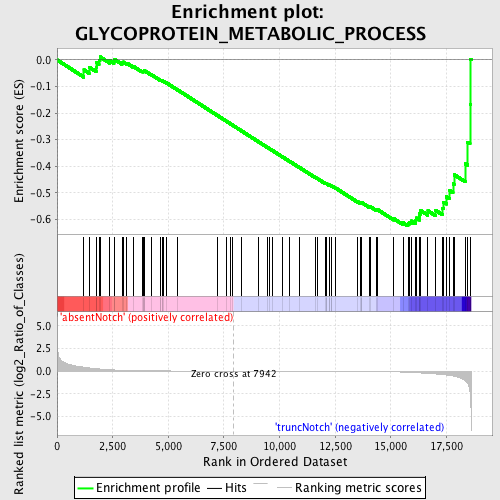
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | GLYCOPROTEIN_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.62492156 |
| Normalized Enrichment Score (NES) | -1.4452047 |
| Nominal p-value | 0.0144665465 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COG3 | 3840746 | 1207 | 0.422 | -0.0376 | No | ||
| 2 | OGT | 2360131 4610333 | 1440 | 0.350 | -0.0273 | No | ||
| 3 | STT3A | 580139 | 1754 | 0.267 | -0.0268 | No | ||
| 4 | LARGE | 6940068 | 1763 | 0.266 | -0.0099 | No | ||
| 5 | GALNT2 | 2260279 4060239 | 1884 | 0.236 | -0.0009 | No | ||
| 6 | POMT1 | 2680047 3440040 6660114 | 1928 | 0.227 | 0.0116 | No | ||
| 7 | MAN1A2 | 5700435 5720114 | 2367 | 0.146 | -0.0025 | No | ||
| 8 | TXNDC4 | 450537 | 2558 | 0.122 | -0.0048 | No | ||
| 9 | ABCG1 | 60692 | 2568 | 0.121 | 0.0026 | No | ||
| 10 | ALG8 | 4150497 4780500 6290563 | 2917 | 0.087 | -0.0105 | No | ||
| 11 | BACE2 | 4060170 5690100 6220133 | 2966 | 0.083 | -0.0077 | No | ||
| 12 | FUT8 | 1340068 2340056 | 3140 | 0.070 | -0.0125 | No | ||
| 13 | FUT4 | 2030324 | 3412 | 0.054 | -0.0236 | No | ||
| 14 | GCS1 | 1740603 | 3820 | 0.035 | -0.0433 | No | ||
| 15 | MAN1C1 | 730133 870398 2340044 | 3862 | 0.033 | -0.0433 | No | ||
| 16 | GAL3ST1 | 5080746 | 3878 | 0.033 | -0.0420 | No | ||
| 17 | DPM1 | 1240050 4810019 6380398 | 3907 | 0.032 | -0.0415 | No | ||
| 18 | ALG2 | 3990091 | 3916 | 0.032 | -0.0398 | No | ||
| 19 | MGAT4A | 5690471 | 4226 | 0.023 | -0.0550 | No | ||
| 20 | NGLY1 | 1570037 | 4653 | 0.016 | -0.0770 | No | ||
| 21 | NCSTN | 940601 6400594 | 4664 | 0.015 | -0.0765 | No | ||
| 22 | RPN1 | 1240113 | 4720 | 0.015 | -0.0785 | No | ||
| 23 | B3GNT5 | 4780528 | 4754 | 0.014 | -0.0794 | No | ||
| 24 | MGEA5 | 6100014 | 4786 | 0.014 | -0.0802 | No | ||
| 25 | B3GALNT1 | 3390746 60131 | 4938 | 0.012 | -0.0875 | No | ||
| 26 | ST8SIA3 | 6940427 | 5430 | 0.009 | -0.1134 | No | ||
| 27 | GALNT7 | 3990280 | 7222 | 0.002 | -0.2099 | No | ||
| 28 | TSPAN8 | 1740538 | 7624 | 0.001 | -0.2315 | No | ||
| 29 | ACHE | 5290750 | 7773 | 0.000 | -0.2395 | No | ||
| 30 | FUT1 | 1500068 | 7862 | 0.000 | -0.2442 | No | ||
| 31 | ADAMTS9 | 2760079 | 8292 | -0.001 | -0.2673 | No | ||
| 32 | B3GALT2 | 430136 | 9061 | -0.003 | -0.3085 | No | ||
| 33 | APOA4 | 4120451 | 9443 | -0.004 | -0.3288 | No | ||
| 34 | ALK | 730008 | 9546 | -0.004 | -0.3341 | No | ||
| 35 | ALG6 | 4060131 | 9658 | -0.004 | -0.3398 | No | ||
| 36 | TUSC3 | 1690411 | 10139 | -0.006 | -0.3653 | No | ||
| 37 | AGA | 6350292 | 10430 | -0.007 | -0.3805 | No | ||
| 38 | FUT9 | 3940121 | 10907 | -0.009 | -0.4056 | No | ||
| 39 | COG7 | 360133 | 11592 | -0.012 | -0.4417 | No | ||
| 40 | TM4SF4 | 540600 | 11685 | -0.012 | -0.4459 | No | ||
| 41 | LRP2 | 1400204 3290706 | 12076 | -0.014 | -0.4660 | No | ||
| 42 | DPM2 | 3610253 | 12092 | -0.015 | -0.4658 | No | ||
| 43 | FUT10 | 6510603 | 12102 | -0.015 | -0.4654 | No | ||
| 44 | B3GALT5 | 4060047 | 12233 | -0.016 | -0.4714 | No | ||
| 45 | LIPC | 2690497 | 12235 | -0.016 | -0.4704 | No | ||
| 46 | APH1A | 4060273 4670465 6450711 | 12343 | -0.017 | -0.4751 | No | ||
| 47 | GCNT3 | 6290239 | 12348 | -0.017 | -0.4742 | No | ||
| 48 | GALNT1 | 4070487 6130100 6250132 6900048 | 12527 | -0.018 | -0.4826 | No | ||
| 49 | FUT2 | 3990047 | 13519 | -0.030 | -0.5341 | No | ||
| 50 | ST8SIA2 | 2680082 | 13642 | -0.032 | -0.5386 | No | ||
| 51 | TSPAN7 | 870133 | 13668 | -0.033 | -0.5378 | No | ||
| 52 | FUT7 | 2900736 | 13669 | -0.033 | -0.5356 | No | ||
| 53 | IL17A | 4280369 | 14028 | -0.040 | -0.5524 | No | ||
| 54 | KEL | 6940440 | 14080 | -0.041 | -0.5524 | No | ||
| 55 | ST8SIA4 | 2680605 3060215 | 14376 | -0.049 | -0.5652 | No | ||
| 56 | GCNT1 | 940129 1780215 2370253 | 14394 | -0.049 | -0.5629 | No | ||
| 57 | LDLR | 5670386 | 15131 | -0.079 | -0.5974 | No | ||
| 58 | CLN3 | 780368 | 15549 | -0.108 | -0.6129 | No | ||
| 59 | POFUT1 | 1570458 | 15773 | -0.129 | -0.6165 | Yes | ||
| 60 | GALNT6 | 3710377 | 15819 | -0.134 | -0.6102 | Yes | ||
| 61 | MAN2B1 | 4210446 | 15918 | -0.145 | -0.6060 | Yes | ||
| 62 | TM4SF5 | 3120280 | 16124 | -0.174 | -0.6057 | Yes | ||
| 63 | ALG1 | 2570112 | 16132 | -0.175 | -0.5947 | Yes | ||
| 64 | COG2 | 5900129 | 16282 | -0.196 | -0.5900 | Yes | ||
| 65 | DOLPP1 | 4780619 6400053 | 16299 | -0.198 | -0.5779 | Yes | ||
| 66 | MGAT4B | 130053 6840100 | 16345 | -0.206 | -0.5669 | Yes | ||
| 67 | B3GALT4 | 2340239 | 16669 | -0.251 | -0.5680 | Yes | ||
| 68 | ALG12 | 1400692 | 17014 | -0.311 | -0.5663 | Yes | ||
| 69 | SDF2 | 2690601 | 17318 | -0.375 | -0.5582 | Yes | ||
| 70 | BACE1 | 2650438 | 17364 | -0.386 | -0.5354 | Yes | ||
| 71 | DPM3 | 50039 | 17494 | -0.423 | -0.5148 | Yes | ||
| 72 | CD37 | 770273 | 17648 | -0.474 | -0.4922 | Yes | ||
| 73 | MPDU1 | 1850091 | 17796 | -0.532 | -0.4654 | Yes | ||
| 74 | PSEN2 | 130382 | 17846 | -0.558 | -0.4317 | Yes | ||
| 75 | GYPC | 2320102 | 18345 | -1.043 | -0.3905 | Yes | ||
| 76 | ALG5 | 3060692 4780093 6590097 | 18451 | -1.323 | -0.3100 | Yes | ||
| 77 | TNIP1 | 2510279 5900041 6510435 | 18560 | -2.265 | -0.1682 | Yes | ||
| 78 | PSEN1 | 130403 2030647 6100603 | 18581 | -2.626 | 0.0019 | Yes |

