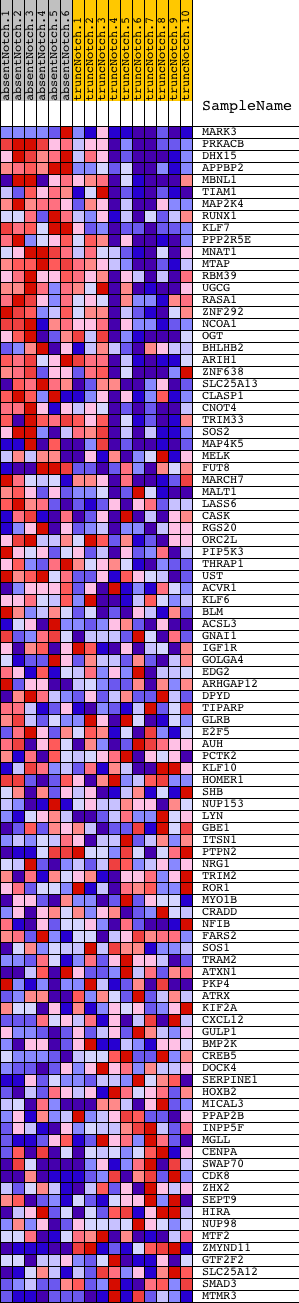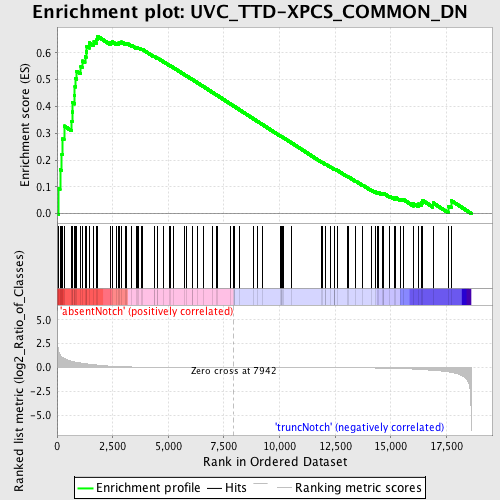
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | UVC_TTD-XPCS_COMMON_DN |
| Enrichment Score (ES) | 0.6625756 |
| Normalized Enrichment Score (NES) | 1.653015 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07341939 |
| FWER p-Value | 0.524 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MARK3 | 2370450 | 61 | 1.732 | 0.0929 | Yes | ||
| 2 | PRKACB | 4210170 | 131 | 1.364 | 0.1648 | Yes | ||
| 3 | DHX15 | 870632 | 212 | 1.110 | 0.2221 | Yes | ||
| 4 | APPBP2 | 5130215 | 240 | 1.048 | 0.2788 | Yes | ||
| 5 | MBNL1 | 2640762 7100048 | 313 | 0.941 | 0.3272 | Yes | ||
| 6 | TIAM1 | 5420288 | 659 | 0.655 | 0.3449 | Yes | ||
| 7 | MAP2K4 | 5130133 | 679 | 0.644 | 0.3796 | Yes | ||
| 8 | RUNX1 | 3840711 | 697 | 0.634 | 0.4139 | Yes | ||
| 9 | KLF7 | 50390 | 787 | 0.591 | 0.4419 | Yes | ||
| 10 | PPP2R5E | 70671 2120056 | 801 | 0.583 | 0.4735 | Yes | ||
| 11 | MNAT1 | 730292 2350364 | 823 | 0.572 | 0.5042 | Yes | ||
| 12 | MTAP | 2120136 | 880 | 0.551 | 0.5317 | Yes | ||
| 13 | RBM39 | 4230458 6450280 | 1067 | 0.471 | 0.5478 | Yes | ||
| 14 | UGCG | 3420053 | 1126 | 0.452 | 0.5698 | Yes | ||
| 15 | RASA1 | 1240315 | 1254 | 0.406 | 0.5855 | Yes | ||
| 16 | ZNF292 | 840736 1500390 | 1303 | 0.388 | 0.6044 | Yes | ||
| 17 | NCOA1 | 3610438 | 1335 | 0.377 | 0.6236 | Yes | ||
| 18 | OGT | 2360131 4610333 | 1440 | 0.350 | 0.6374 | Yes | ||
| 19 | BHLHB2 | 7040603 | 1656 | 0.290 | 0.6419 | Yes | ||
| 20 | ARIH1 | 6900025 | 1787 | 0.260 | 0.6494 | Yes | ||
| 21 | ZNF638 | 6510112 | 1806 | 0.256 | 0.6626 | Yes | ||
| 22 | SLC25A13 | 4920072 5690722 | 2404 | 0.142 | 0.6382 | No | ||
| 23 | CLASP1 | 6860279 | 2482 | 0.130 | 0.6412 | No | ||
| 24 | CNOT4 | 2320242 6760706 | 2689 | 0.107 | 0.6361 | No | ||
| 25 | TRIM33 | 580619 2230280 3990433 6200747 | 2775 | 0.099 | 0.6370 | No | ||
| 26 | SOS2 | 2060722 | 2821 | 0.096 | 0.6399 | No | ||
| 27 | MAP4K5 | 3190286 4760300 5270070 5550494 | 2896 | 0.089 | 0.6408 | No | ||
| 28 | MELK | 130022 5220537 | 3066 | 0.075 | 0.6359 | No | ||
| 29 | FUT8 | 1340068 2340056 | 3140 | 0.070 | 0.6358 | No | ||
| 30 | MARCH7 | 3800020 6620168 | 3348 | 0.057 | 0.6279 | No | ||
| 31 | MALT1 | 4670292 | 3555 | 0.047 | 0.6193 | No | ||
| 32 | LASS6 | 5890576 6520138 | 3607 | 0.044 | 0.6190 | No | ||
| 33 | CASK | 2340215 3290576 6770215 | 3677 | 0.041 | 0.6175 | No | ||
| 34 | RGS20 | 4540541 4730086 | 3782 | 0.036 | 0.6139 | No | ||
| 35 | ORC2L | 1990470 6510019 | 3821 | 0.035 | 0.6138 | No | ||
| 36 | PIP5K3 | 5360112 | 4356 | 0.020 | 0.5861 | No | ||
| 37 | THRAP1 | 2470121 | 4382 | 0.020 | 0.5858 | No | ||
| 38 | UST | 2100053 | 4492 | 0.018 | 0.5809 | No | ||
| 39 | ACVR1 | 6840671 | 4525 | 0.017 | 0.5801 | No | ||
| 40 | KLF6 | 110541 5360026 | 4800 | 0.014 | 0.5661 | No | ||
| 41 | BLM | 520619 5570170 | 5053 | 0.011 | 0.5531 | No | ||
| 42 | ACSL3 | 3140195 | 5118 | 0.011 | 0.5502 | No | ||
| 43 | GNAI1 | 4560390 | 5244 | 0.010 | 0.5440 | No | ||
| 44 | IGF1R | 3360494 | 5733 | 0.007 | 0.5181 | No | ||
| 45 | GOLGA4 | 460300 | 5831 | 0.007 | 0.5132 | No | ||
| 46 | EDG2 | 6290215 | 6071 | 0.006 | 0.5006 | No | ||
| 47 | ARHGAP12 | 1050181 | 6072 | 0.006 | 0.5009 | No | ||
| 48 | DPYD | 6110095 | 6321 | 0.005 | 0.4878 | No | ||
| 49 | TIPARP | 50397 | 6570 | 0.004 | 0.4746 | No | ||
| 50 | GLRB | 2510433 4280736 | 7003 | 0.002 | 0.4514 | No | ||
| 51 | E2F5 | 5860575 | 7165 | 0.002 | 0.4428 | No | ||
| 52 | AUH | 5570152 | 7201 | 0.002 | 0.4411 | No | ||
| 53 | PCTK2 | 5700673 | 7793 | 0.000 | 0.4092 | No | ||
| 54 | KLF10 | 4850056 | 7935 | 0.000 | 0.4016 | No | ||
| 55 | HOMER1 | 360131 1690092 1990300 4590068 | 7976 | -0.000 | 0.3994 | No | ||
| 56 | SHB | 4920494 | 7988 | -0.000 | 0.3988 | No | ||
| 57 | NUP153 | 7000452 | 8185 | -0.001 | 0.3883 | No | ||
| 58 | LYN | 6040600 | 8811 | -0.002 | 0.3546 | No | ||
| 59 | GBE1 | 2190368 | 9006 | -0.003 | 0.3443 | No | ||
| 60 | ITSN1 | 2640577 | 9236 | -0.003 | 0.3321 | No | ||
| 61 | PTPN2 | 130315 | 10041 | -0.006 | 0.2890 | No | ||
| 62 | NRG1 | 1050332 | 10073 | -0.006 | 0.2877 | No | ||
| 63 | TRIM2 | 430546 1170286 | 10084 | -0.006 | 0.2874 | No | ||
| 64 | ROR1 | 6220026 | 10138 | -0.006 | 0.2849 | No | ||
| 65 | MYO1B | 770372 | 10156 | -0.006 | 0.2843 | No | ||
| 66 | CRADD | 6770520 | 10529 | -0.007 | 0.2646 | No | ||
| 67 | NFIB | 460450 | 11897 | -0.013 | 0.1916 | No | ||
| 68 | FARS2 | 5340575 | 11924 | -0.013 | 0.1909 | No | ||
| 69 | SOS1 | 7050338 | 12053 | -0.014 | 0.1848 | No | ||
| 70 | TRAM2 | 3940112 | 12079 | -0.015 | 0.1843 | No | ||
| 71 | ATXN1 | 5550156 | 12306 | -0.016 | 0.1730 | No | ||
| 72 | PKP4 | 3710400 5890097 | 12452 | -0.017 | 0.1661 | No | ||
| 73 | ATRX | 2340039 3610132 4210373 | 12464 | -0.018 | 0.1665 | No | ||
| 74 | KIF2A | 3990286 6130575 | 12485 | -0.018 | 0.1664 | No | ||
| 75 | CXCL12 | 580546 4150750 4570068 | 12619 | -0.019 | 0.1602 | No | ||
| 76 | GULP1 | 2230725 2350039 5570446 | 13033 | -0.023 | 0.1392 | No | ||
| 77 | BMP2K | 60541 5570504 | 13119 | -0.024 | 0.1360 | No | ||
| 78 | CREB5 | 2320368 | 13431 | -0.028 | 0.1208 | No | ||
| 79 | DOCK4 | 5910102 | 13732 | -0.034 | 0.1065 | No | ||
| 80 | SERPINE1 | 4210403 | 14127 | -0.042 | 0.0875 | No | ||
| 81 | HOXB2 | 6450592 | 14288 | -0.047 | 0.0815 | No | ||
| 82 | MICAL3 | 6040333 | 14390 | -0.049 | 0.0788 | No | ||
| 83 | PPAP2B | 3190397 4730280 | 14423 | -0.050 | 0.0798 | No | ||
| 84 | INPP5F | 60010 5360164 | 14615 | -0.056 | 0.0726 | No | ||
| 85 | MGLL | 2030446 4210520 | 14659 | -0.058 | 0.0735 | No | ||
| 86 | CENPA | 5080154 | 14675 | -0.058 | 0.0759 | No | ||
| 87 | SWAP70 | 2810577 2850739 5130594 | 14943 | -0.069 | 0.0653 | No | ||
| 88 | CDK8 | 1050113 1190601 1780358 6110102 | 15161 | -0.081 | 0.0581 | No | ||
| 89 | ZHX2 | 2900452 | 15222 | -0.084 | 0.0595 | No | ||
| 90 | SEPT9 | 1940040 | 15438 | -0.098 | 0.0534 | No | ||
| 91 | HIRA | 4480609 5270167 | 15548 | -0.108 | 0.0534 | No | ||
| 92 | NUP98 | 2850465 | 16018 | -0.162 | 0.0371 | No | ||
| 93 | MTF2 | 50333 | 16233 | -0.189 | 0.0360 | No | ||
| 94 | ZMYND11 | 630181 2570019 4850138 5360040 | 16370 | -0.211 | 0.0404 | No | ||
| 95 | GTF2F2 | 2690546 | 16408 | -0.216 | 0.0503 | No | ||
| 96 | SLC25A12 | 7400731 | 16895 | -0.290 | 0.0402 | No | ||
| 97 | SMAD3 | 6450671 | 17586 | -0.454 | 0.0281 | No | ||
| 98 | MTMR3 | 1050014 4920180 | 17716 | -0.495 | 0.0486 | No |