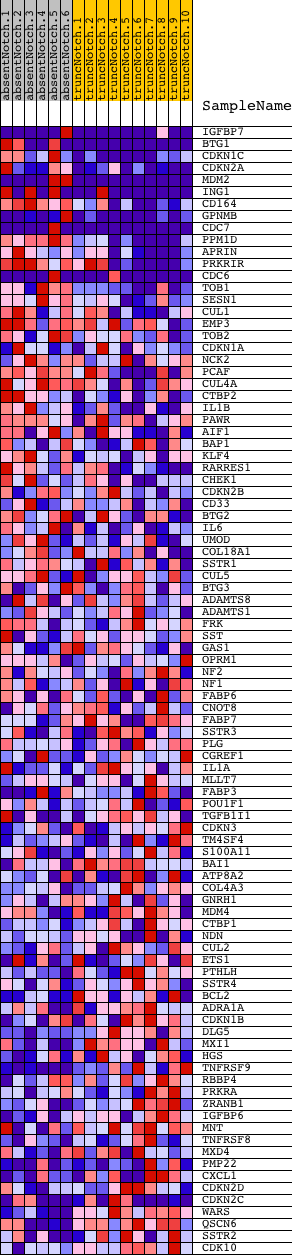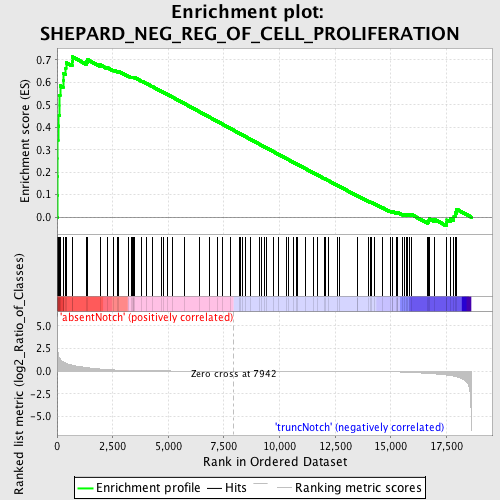
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | SHEPARD_NEG_REG_OF_CELL_PROLIFERATION |
| Enrichment Score (ES) | 0.7137976 |
| Normalized Enrichment Score (NES) | 1.7376375 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.029574601 |
| FWER p-Value | 0.074 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IGFBP7 | 520411 3060110 5290152 | 9 | 3.084 | 0.0981 | Yes | ||
| 2 | BTG1 | 4200735 6040131 6200133 | 14 | 2.662 | 0.1830 | Yes | ||
| 3 | CDKN1C | 6520577 | 20 | 2.517 | 0.2632 | Yes | ||
| 4 | CDKN2A | 4670215 4760047 | 22 | 2.420 | 0.3405 | Yes | ||
| 5 | MDM2 | 3450053 5080138 | 41 | 2.078 | 0.4060 | Yes | ||
| 6 | ING1 | 5690010 6520056 | 81 | 1.566 | 0.4540 | Yes | ||
| 7 | CD164 | 3830594 | 112 | 1.406 | 0.4973 | Yes | ||
| 8 | GPNMB | 1780138 2340176 3390403 4560156 | 113 | 1.401 | 0.5421 | Yes | ||
| 9 | CDC7 | 4060546 4850041 | 133 | 1.359 | 0.5845 | Yes | ||
| 10 | PPM1D | 1690193 | 270 | 1.000 | 0.6092 | Yes | ||
| 11 | APRIN | 2810050 | 277 | 0.987 | 0.6404 | Yes | ||
| 12 | PRKRIR | 2370402 | 389 | 0.856 | 0.6618 | Yes | ||
| 13 | CDC6 | 4570296 5360600 | 400 | 0.848 | 0.6883 | Yes | ||
| 14 | TOB1 | 4150138 | 675 | 0.646 | 0.6942 | Yes | ||
| 15 | SESN1 | 2120021 | 691 | 0.638 | 0.7138 | Yes | ||
| 16 | CUL1 | 1990632 | 1308 | 0.386 | 0.6929 | No | ||
| 17 | EMP3 | 1500722 1570039 | 1378 | 0.365 | 0.7008 | No | ||
| 18 | TOB2 | 1240465 | 1933 | 0.225 | 0.6781 | No | ||
| 19 | CDKN1A | 4050088 6400706 | 2242 | 0.164 | 0.6667 | No | ||
| 20 | NCK2 | 2510010 | 2539 | 0.124 | 0.6547 | No | ||
| 21 | PCAF | 2230161 2570369 6550451 | 2700 | 0.107 | 0.6495 | No | ||
| 22 | CUL4A | 1170088 2320008 2470278 | 2752 | 0.101 | 0.6500 | No | ||
| 23 | CTBP2 | 430309 3710079 | 3199 | 0.067 | 0.6280 | No | ||
| 24 | IL1B | 2640364 | 3322 | 0.059 | 0.6233 | No | ||
| 25 | PAWR | 5130546 6330706 | 3379 | 0.056 | 0.6221 | No | ||
| 26 | AIF1 | 130332 5220017 | 3415 | 0.053 | 0.6219 | No | ||
| 27 | BAP1 | 3830131 | 3444 | 0.052 | 0.6221 | No | ||
| 28 | KLF4 | 1850022 3830239 5570750 | 3493 | 0.050 | 0.6211 | No | ||
| 29 | RARRES1 | 770139 5860390 | 3801 | 0.035 | 0.6056 | No | ||
| 30 | CHEK1 | 50167 1940300 | 4014 | 0.028 | 0.5951 | No | ||
| 31 | CDKN2B | 6020040 | 4293 | 0.021 | 0.5807 | No | ||
| 32 | CD33 | 3990735 | 4694 | 0.015 | 0.5596 | No | ||
| 33 | BTG2 | 2350411 | 4794 | 0.014 | 0.5547 | No | ||
| 34 | IL6 | 380133 | 4975 | 0.012 | 0.5454 | No | ||
| 35 | UMOD | 1980603 | 5175 | 0.010 | 0.5350 | No | ||
| 36 | COL18A1 | 610301 4570338 | 5714 | 0.007 | 0.5061 | No | ||
| 37 | SSTR1 | 2630471 | 6414 | 0.004 | 0.4685 | No | ||
| 38 | CUL5 | 450142 | 6869 | 0.003 | 0.4441 | No | ||
| 39 | BTG3 | 7050079 | 7195 | 0.002 | 0.4266 | No | ||
| 40 | ADAMTS8 | 3990497 | 7443 | 0.001 | 0.4134 | No | ||
| 41 | ADAMTS1 | 3140286 5860463 | 7806 | 0.000 | 0.3938 | No | ||
| 42 | FRK | 5130333 | 8206 | -0.001 | 0.3723 | No | ||
| 43 | SST | 6590142 | 8260 | -0.001 | 0.3695 | No | ||
| 44 | GAS1 | 2120504 | 8332 | -0.001 | 0.3657 | No | ||
| 45 | OPRM1 | 5360279 | 8473 | -0.001 | 0.3581 | No | ||
| 46 | NF2 | 4150735 6450139 | 8710 | -0.002 | 0.3455 | No | ||
| 47 | NF1 | 6980433 | 9083 | -0.003 | 0.3255 | No | ||
| 48 | FABP6 | 7000278 | 9173 | -0.003 | 0.3208 | No | ||
| 49 | CNOT8 | 4590129 | 9323 | -0.003 | 0.3128 | No | ||
| 50 | FABP7 | 4590142 | 9433 | -0.004 | 0.3071 | No | ||
| 51 | SSTR3 | 5420064 | 9714 | -0.005 | 0.2921 | No | ||
| 52 | PLG | 3360270 3840100 | 9939 | -0.005 | 0.2802 | No | ||
| 53 | CGREF1 | 4540546 | 10300 | -0.006 | 0.2609 | No | ||
| 54 | IL1A | 3610056 | 10395 | -0.007 | 0.2561 | No | ||
| 55 | MLLT7 | 4480707 | 10618 | -0.008 | 0.2443 | No | ||
| 56 | FABP3 | 6860452 | 10761 | -0.008 | 0.2369 | No | ||
| 57 | POU1F1 | 450148 | 10807 | -0.008 | 0.2348 | No | ||
| 58 | TGFB1I1 | 2060288 6550450 | 11151 | -0.009 | 0.2166 | No | ||
| 59 | CDKN3 | 110520 | 11537 | -0.011 | 0.1961 | No | ||
| 60 | TM4SF4 | 540600 | 11685 | -0.012 | 0.1886 | No | ||
| 61 | S100A11 | 2260064 | 12006 | -0.014 | 0.1718 | No | ||
| 62 | BAI1 | 6350053 | 12041 | -0.014 | 0.1704 | No | ||
| 63 | ATP8A2 | 1580086 3940025 | 12207 | -0.015 | 0.1620 | No | ||
| 64 | COL4A3 | 5910075 | 12595 | -0.019 | 0.1417 | No | ||
| 65 | GNRH1 | 430167 | 12670 | -0.019 | 0.1383 | No | ||
| 66 | MDM4 | 4070504 4780008 | 13496 | -0.030 | 0.0947 | No | ||
| 67 | CTBP1 | 3780315 | 13517 | -0.030 | 0.0946 | No | ||
| 68 | NDN | 5670075 | 13977 | -0.039 | 0.0710 | No | ||
| 69 | CUL2 | 4200278 | 14066 | -0.041 | 0.0676 | No | ||
| 70 | ETS1 | 5270278 6450717 6620465 | 14113 | -0.042 | 0.0664 | No | ||
| 71 | PTHLH | 5290739 | 14267 | -0.046 | 0.0597 | No | ||
| 72 | SSTR4 | 6100398 | 14624 | -0.056 | 0.0422 | No | ||
| 73 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 14977 | -0.070 | 0.0255 | No | ||
| 74 | ADRA1A | 1230446 2260390 | 15057 | -0.075 | 0.0236 | No | ||
| 75 | CDKN1B | 3800025 6450044 | 15072 | -0.076 | 0.0253 | No | ||
| 76 | DLG5 | 450215 | 15235 | -0.084 | 0.0192 | No | ||
| 77 | MXI1 | 5050064 5130484 | 15279 | -0.087 | 0.0197 | No | ||
| 78 | HGS | 5570722 | 15311 | -0.089 | 0.0209 | No | ||
| 79 | TNFRSF9 | 2510400 6650484 | 15523 | -0.105 | 0.0128 | No | ||
| 80 | RBBP4 | 6650528 | 15602 | -0.113 | 0.0122 | No | ||
| 81 | PRKRA | 2510600 | 15688 | -0.121 | 0.0115 | No | ||
| 82 | ZRANB1 | 510373 4540278 | 15755 | -0.128 | 0.0120 | No | ||
| 83 | IGFBP6 | 1580181 | 15840 | -0.136 | 0.0118 | No | ||
| 84 | MNT | 4920575 | 15919 | -0.145 | 0.0123 | No | ||
| 85 | TNFRSF8 | 870136 4810446 | 16652 | -0.248 | -0.0193 | No | ||
| 86 | MXD4 | 4810113 | 16709 | -0.257 | -0.0141 | No | ||
| 87 | PMP22 | 4010239 6550072 | 16723 | -0.259 | -0.0065 | No | ||
| 88 | CXCL1 | 2690537 | 16950 | -0.300 | -0.0092 | No | ||
| 89 | CDKN2D | 6040035 | 17507 | -0.427 | -0.0255 | No | ||
| 90 | CDKN2C | 5050750 5130148 | 17523 | -0.433 | -0.0125 | No | ||
| 91 | WARS | 2570717 3290139 | 17685 | -0.486 | -0.0057 | No | ||
| 92 | QSCN6 | 3120441 | 17837 | -0.554 | 0.0039 | No | ||
| 93 | SSTR2 | 4590687 | 17892 | -0.581 | 0.0195 | No | ||
| 94 | CDK10 | 60164 1190717 3130128 3990044 | 17944 | -0.610 | 0.0363 | No |