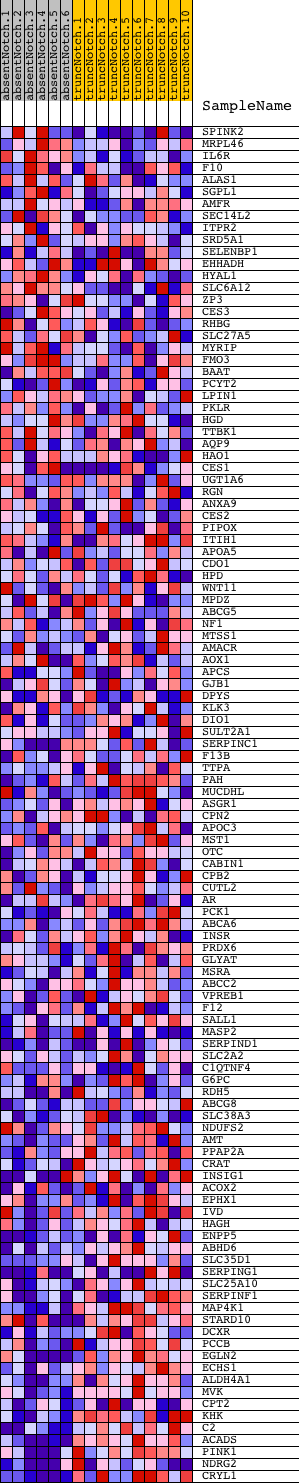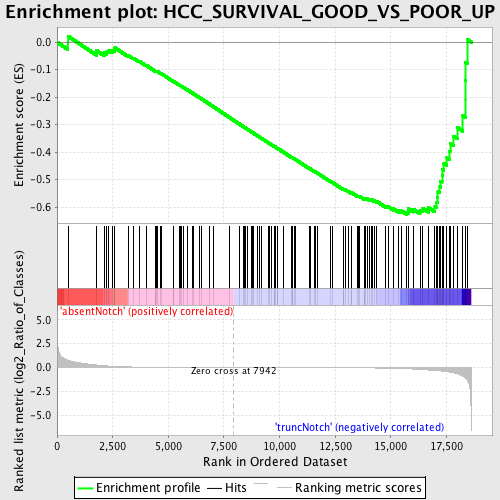
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | HCC_SURVIVAL_GOOD_VS_POOR_UP |
| Enrichment Score (ES) | -0.62652194 |
| Normalized Enrichment Score (NES) | -1.5285501 |
| Nominal p-value | 0.001934236 |
| FDR q-value | 0.4683475 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SPINK2 | 3990093 | 489 | 0.766 | 0.0220 | No | ||
| 2 | MRPL46 | 360736 | 1777 | 0.263 | -0.0309 | No | ||
| 3 | IL6R | 520706 3800215 | 2135 | 0.185 | -0.0385 | No | ||
| 4 | F10 | 3830450 | 2239 | 0.164 | -0.0337 | No | ||
| 5 | ALAS1 | 6400440 | 2329 | 0.151 | -0.0290 | No | ||
| 6 | SGPL1 | 4480059 | 2508 | 0.127 | -0.0306 | No | ||
| 7 | AMFR | 2810041 | 2595 | 0.119 | -0.0277 | No | ||
| 8 | SEC14L2 | 2640370 | 2600 | 0.119 | -0.0204 | No | ||
| 9 | ITPR2 | 5360128 | 3200 | 0.067 | -0.0486 | No | ||
| 10 | SRD5A1 | 5910347 | 3433 | 0.053 | -0.0578 | No | ||
| 11 | SELENBP1 | 3850575 | 3695 | 0.040 | -0.0694 | No | ||
| 12 | EHHADH | 6200315 | 4004 | 0.029 | -0.0842 | No | ||
| 13 | HYAL1 | 3850341 | 4421 | 0.019 | -0.1055 | No | ||
| 14 | SLC6A12 | 3170685 | 4442 | 0.018 | -0.1054 | No | ||
| 15 | ZP3 | 4060767 | 4464 | 0.018 | -0.1054 | No | ||
| 16 | CES3 | 5050167 3290458 | 4506 | 0.018 | -0.1065 | No | ||
| 17 | RHBG | 2810736 | 4642 | 0.016 | -0.1128 | No | ||
| 18 | SLC27A5 | 2850717 | 4699 | 0.015 | -0.1149 | No | ||
| 19 | MYRIP | 1580471 | 5223 | 0.010 | -0.1426 | No | ||
| 20 | FMO3 | 2480369 | 5230 | 0.010 | -0.1423 | No | ||
| 21 | BAAT | 730739 | 5493 | 0.008 | -0.1559 | No | ||
| 22 | PCYT2 | 2630092 4590278 | 5531 | 0.008 | -0.1574 | No | ||
| 23 | LPIN1 | 1770358 3120059 | 5587 | 0.008 | -0.1599 | No | ||
| 24 | PKLR | 1170400 2470114 | 5677 | 0.007 | -0.1642 | No | ||
| 25 | HGD | 460390 | 5864 | 0.007 | -0.1738 | No | ||
| 26 | TTBK1 | 60605 1850722 | 6070 | 0.006 | -0.1846 | No | ||
| 27 | AQP9 | 2650047 | 6139 | 0.005 | -0.1879 | No | ||
| 28 | HAO1 | 7050093 | 6389 | 0.004 | -0.2011 | No | ||
| 29 | CES1 | 1400494 | 6471 | 0.004 | -0.2052 | No | ||
| 30 | UGT1A6 | 4610368 6900086 4540092 2630075 | 6827 | 0.003 | -0.2242 | No | ||
| 31 | RGN | 6200095 | 7045 | 0.002 | -0.2358 | No | ||
| 32 | ANXA9 | 1190195 | 7742 | 0.000 | -0.2733 | No | ||
| 33 | CES2 | 5340609 | 8202 | -0.001 | -0.2981 | No | ||
| 34 | PIPOX | 70750 | 8361 | -0.001 | -0.3066 | No | ||
| 35 | ITIH1 | 670044 | 8436 | -0.001 | -0.3105 | No | ||
| 36 | APOA5 | 6130471 | 8445 | -0.001 | -0.3109 | No | ||
| 37 | CDO1 | 2480279 | 8579 | -0.002 | -0.3179 | No | ||
| 38 | HPD | 3120519 | 8741 | -0.002 | -0.3265 | No | ||
| 39 | WNT11 | 1230278 | 8794 | -0.002 | -0.3292 | No | ||
| 40 | MPDZ | 130026 1990064 4050577 | 8830 | -0.002 | -0.3309 | No | ||
| 41 | ABCG5 | 2810670 | 9004 | -0.003 | -0.3401 | No | ||
| 42 | NF1 | 6980433 | 9083 | -0.003 | -0.3442 | No | ||
| 43 | MTSS1 | 780435 2370114 | 9195 | -0.003 | -0.3500 | No | ||
| 44 | AMACR | 780215 | 9481 | -0.004 | -0.3651 | No | ||
| 45 | AOX1 | 110082 6290450 | 9524 | -0.004 | -0.3671 | No | ||
| 46 | APCS | 2060500 | 9617 | -0.004 | -0.3718 | No | ||
| 47 | GJB1 | 1240441 | 9765 | -0.005 | -0.3795 | No | ||
| 48 | DPYS | 3130670 | 9787 | -0.005 | -0.3803 | No | ||
| 49 | KLK3 | 1500278 | 9801 | -0.005 | -0.3807 | No | ||
| 50 | DIO1 | 4570279 5290112 5420148 | 9891 | -0.005 | -0.3852 | No | ||
| 51 | SULT2A1 | 2650286 | 9912 | -0.005 | -0.3859 | No | ||
| 52 | SERPINC1 | 1780717 | 10176 | -0.006 | -0.3998 | No | ||
| 53 | F13B | 6220278 | 10534 | -0.007 | -0.4186 | No | ||
| 54 | TTPA | 630576 | 10567 | -0.007 | -0.4199 | No | ||
| 55 | PAH | 3800309 | 10568 | -0.007 | -0.4194 | No | ||
| 56 | MUCDHL | 5720056 | 10669 | -0.008 | -0.4243 | No | ||
| 57 | ASGR1 | 1850088 | 10712 | -0.008 | -0.4261 | No | ||
| 58 | CPN2 | 6590291 | 11333 | -0.010 | -0.4589 | No | ||
| 59 | APOC3 | 2030168 | 11381 | -0.011 | -0.4608 | No | ||
| 60 | MST1 | 1400403 | 11580 | -0.011 | -0.4708 | No | ||
| 61 | OTC | 1740494 | 11597 | -0.012 | -0.4709 | No | ||
| 62 | CABIN1 | 6900446 | 11721 | -0.012 | -0.4768 | No | ||
| 63 | CPB2 | 4150167 6100333 6590735 | 12278 | -0.016 | -0.5058 | No | ||
| 64 | CUTL2 | 2940692 6220687 | 12382 | -0.017 | -0.5103 | No | ||
| 65 | AR | 6380167 | 12867 | -0.021 | -0.5351 | No | ||
| 66 | PCK1 | 7000358 | 12894 | -0.022 | -0.5351 | No | ||
| 67 | ABCA6 | 130538 1050706 | 12940 | -0.022 | -0.5362 | No | ||
| 68 | INSR | 1190504 | 13090 | -0.024 | -0.5427 | No | ||
| 69 | PRDX6 | 4920397 6380601 | 13223 | -0.025 | -0.5482 | No | ||
| 70 | GLYAT | 6200239 | 13227 | -0.025 | -0.5468 | No | ||
| 71 | MSRA | 4570411 | 13510 | -0.030 | -0.5601 | No | ||
| 72 | ABCC2 | 380202 5390300 | 13558 | -0.031 | -0.5607 | No | ||
| 73 | VPREB1 | 3520280 | 13596 | -0.032 | -0.5607 | No | ||
| 74 | F12 | 1090215 3520736 | 13795 | -0.035 | -0.5692 | No | ||
| 75 | SALL1 | 5420020 7050195 | 13824 | -0.036 | -0.5685 | No | ||
| 76 | MASP2 | 360110 | 13876 | -0.037 | -0.5689 | No | ||
| 77 | SERPIND1 | 6110059 | 13939 | -0.038 | -0.5699 | No | ||
| 78 | SLC2A2 | 5130537 5720722 6770079 | 14041 | -0.040 | -0.5728 | No | ||
| 79 | C1QTNF4 | 60619 | 14109 | -0.042 | -0.5738 | No | ||
| 80 | G6PC | 430093 | 14155 | -0.043 | -0.5735 | No | ||
| 81 | RDH5 | 2970736 3940632 5340730 6370093 4610059 | 14249 | -0.046 | -0.5756 | No | ||
| 82 | ABCG8 | 4780605 | 14371 | -0.049 | -0.5791 | No | ||
| 83 | SLC38A3 | 5340142 | 14770 | -0.061 | -0.5967 | No | ||
| 84 | NDUFS2 | 4850020 6200402 | 14882 | -0.066 | -0.5986 | No | ||
| 85 | AMT | 3180450 | 15103 | -0.077 | -0.6056 | No | ||
| 86 | PPAP2A | 1170397 3440288 3930270 | 15357 | -0.092 | -0.6134 | No | ||
| 87 | CRAT | 540020 2060364 3520148 | 15478 | -0.101 | -0.6135 | No | ||
| 88 | INSIG1 | 1500332 3450458 4540301 | 15720 | -0.125 | -0.6186 | Yes | ||
| 89 | ACOX2 | 450017 | 15787 | -0.131 | -0.6139 | Yes | ||
| 90 | EPHX1 | 1740136 | 15789 | -0.131 | -0.6057 | Yes | ||
| 91 | IVD | 2510673 | 16009 | -0.160 | -0.6074 | Yes | ||
| 92 | HAGH | 540358 | 16313 | -0.200 | -0.6111 | Yes | ||
| 93 | ENPP5 | 1780338 | 16420 | -0.218 | -0.6031 | Yes | ||
| 94 | ABHD6 | 630717 | 16693 | -0.255 | -0.6017 | Yes | ||
| 95 | SLC35D1 | 1740520 | 16983 | -0.306 | -0.5980 | Yes | ||
| 96 | SERPING1 | 5550440 | 17069 | -0.321 | -0.5822 | Yes | ||
| 97 | SLC25A10 | 1400609 | 17099 | -0.326 | -0.5632 | Yes | ||
| 98 | SERPINF1 | 7040367 | 17119 | -0.329 | -0.5434 | Yes | ||
| 99 | MAP4K1 | 4060692 | 17206 | -0.350 | -0.5260 | Yes | ||
| 100 | STARD10 | 1400619 4570170 | 17245 | -0.357 | -0.5054 | Yes | ||
| 101 | DCXR | 4730551 | 17302 | -0.371 | -0.4850 | Yes | ||
| 102 | PCCB | 50670 | 17316 | -0.374 | -0.4620 | Yes | ||
| 103 | EGLN2 | 540086 | 17385 | -0.391 | -0.4410 | Yes | ||
| 104 | ECHS1 | 2970184 | 17522 | -0.433 | -0.4210 | Yes | ||
| 105 | ALDH4A1 | 2450450 | 17651 | -0.474 | -0.3979 | Yes | ||
| 106 | MVK | 1450717 | 17673 | -0.481 | -0.3686 | Yes | ||
| 107 | CPT2 | 3940504 6180075 | 17823 | -0.548 | -0.3420 | Yes | ||
| 108 | KHK | 1090204 3870204 | 18007 | -0.646 | -0.3110 | Yes | ||
| 109 | C2 | 5390465 | 18239 | -0.888 | -0.2674 | Yes | ||
| 110 | ACADS | 110746 | 18349 | -1.049 | -0.2070 | Yes | ||
| 111 | PINK1 | 380044 580577 | 18350 | -1.049 | -0.1407 | Yes | ||
| 112 | NDRG2 | 450403 | 18360 | -1.078 | -0.0730 | Yes | ||
| 113 | CRYL1 | 1340427 | 18459 | -1.372 | 0.0085 | Yes |