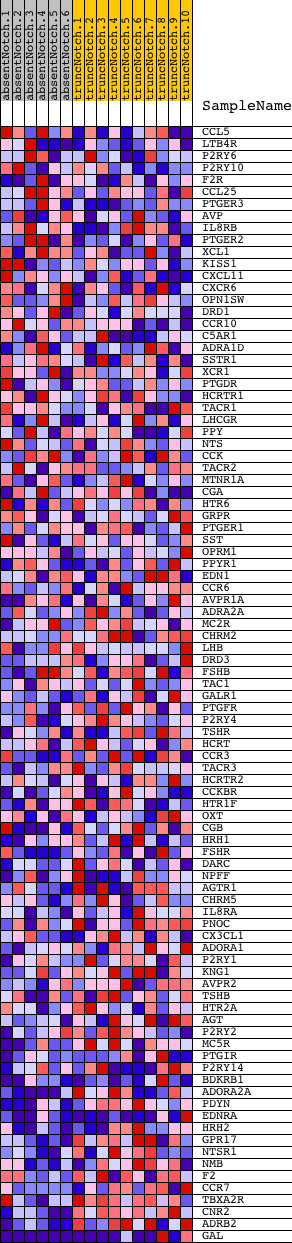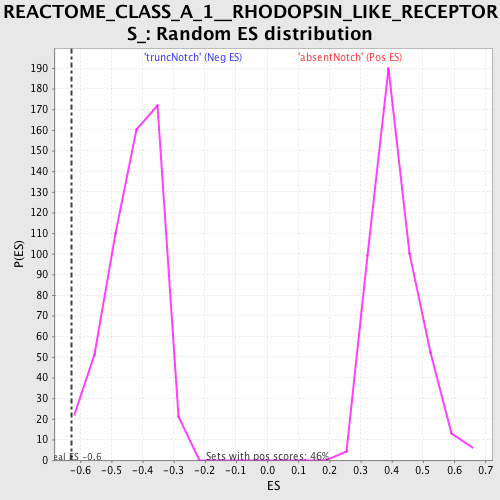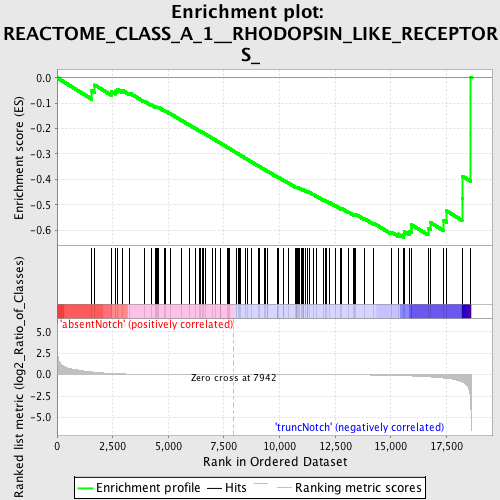
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | truncNotch |
| GeneSet | REACTOME_CLASS_A_1__RHODOPSIN_LIKE_RECEPTORS_ |
| Enrichment Score (ES) | -0.6288667 |
| Normalized Enrichment Score (NES) | -1.4766023 |
| Nominal p-value | 0.009328358 |
| FDR q-value | 0.4652268 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CCL5 | 3710397 | 1562 | 0.321 | -0.0509 | No | ||
| 2 | LTB4R | 1770056 | 1690 | 0.282 | -0.0284 | No | ||
| 3 | P2RY6 | 5290400 | 2435 | 0.137 | -0.0543 | No | ||
| 4 | P2RY10 | 6370039 6840204 | 2616 | 0.117 | -0.0518 | No | ||
| 5 | F2R | 4810180 | 2696 | 0.107 | -0.0450 | No | ||
| 6 | CCL25 | 450541 540435 | 2948 | 0.084 | -0.0497 | No | ||
| 7 | PTGER3 | 2900739 6770750 | 3275 | 0.062 | -0.0609 | No | ||
| 8 | AVP | 2100113 | 3908 | 0.032 | -0.0917 | No | ||
| 9 | IL8RB | 450592 1170537 | 4225 | 0.023 | -0.1064 | No | ||
| 10 | PTGER2 | 3170039 | 4400 | 0.019 | -0.1138 | No | ||
| 11 | XCL1 | 3800504 | 4457 | 0.018 | -0.1149 | No | ||
| 12 | KISS1 | 1580398 | 4505 | 0.018 | -0.1156 | No | ||
| 13 | CXCL11 | 1090551 | 4541 | 0.017 | -0.1157 | No | ||
| 14 | CXCR6 | 3190440 | 4804 | 0.014 | -0.1285 | No | ||
| 15 | OPN1SW | 6420377 | 4893 | 0.013 | -0.1319 | No | ||
| 16 | DRD1 | 430025 | 5090 | 0.011 | -0.1413 | No | ||
| 17 | CCR10 | 4050097 | 5569 | 0.008 | -0.1663 | No | ||
| 18 | C5AR1 | 4540402 | 5928 | 0.006 | -0.1850 | No | ||
| 19 | ADRA1D | 380035 540025 | 6234 | 0.005 | -0.2009 | No | ||
| 20 | SSTR1 | 2630471 | 6414 | 0.004 | -0.2102 | No | ||
| 21 | XCR1 | 5080072 | 6456 | 0.004 | -0.2119 | No | ||
| 22 | PTGDR | 3850161 | 6513 | 0.004 | -0.2145 | No | ||
| 23 | HCRTR1 | 1580273 | 6561 | 0.004 | -0.2167 | No | ||
| 24 | TACR1 | 70358 3840411 | 6667 | 0.004 | -0.2220 | No | ||
| 25 | LHCGR | 4560465 5860551 | 7000 | 0.002 | -0.2396 | No | ||
| 26 | PPY | 2340373 | 7121 | 0.002 | -0.2459 | No | ||
| 27 | NTS | 380300 2120592 | 7341 | 0.002 | -0.2576 | No | ||
| 28 | CCK | 6370368 | 7345 | 0.002 | -0.2576 | No | ||
| 29 | TACR2 | 1740358 | 7671 | 0.001 | -0.2750 | No | ||
| 30 | MTNR1A | 380204 | 7702 | 0.001 | -0.2766 | No | ||
| 31 | CGA | 630403 | 7770 | 0.000 | -0.2802 | No | ||
| 32 | HTR6 | 4850022 | 8052 | -0.000 | -0.2953 | No | ||
| 33 | GRPR | 6020170 | 8154 | -0.000 | -0.3007 | No | ||
| 34 | PTGER1 | 6100132 | 8205 | -0.001 | -0.3033 | No | ||
| 35 | SST | 6590142 | 8260 | -0.001 | -0.3062 | No | ||
| 36 | OPRM1 | 5360279 | 8473 | -0.001 | -0.3175 | No | ||
| 37 | PPYR1 | 1410072 | 8566 | -0.002 | -0.3223 | No | ||
| 38 | EDN1 | 1770047 | 8742 | -0.002 | -0.3315 | No | ||
| 39 | CCR6 | 5720368 6020176 | 9058 | -0.003 | -0.3482 | No | ||
| 40 | AVPR1A | 2120300 | 9113 | -0.003 | -0.3509 | No | ||
| 41 | ADRA2A | 5340520 | 9334 | -0.004 | -0.3624 | No | ||
| 42 | MC2R | 2970504 | 9388 | -0.004 | -0.3649 | No | ||
| 43 | CHRM2 | 870750 | 9448 | -0.004 | -0.3676 | No | ||
| 44 | LHB | 1450368 | 9467 | -0.004 | -0.3682 | No | ||
| 45 | DRD3 | 4780402 | 9893 | -0.005 | -0.3906 | No | ||
| 46 | FSHB | 7100592 | 9908 | -0.005 | -0.3908 | No | ||
| 47 | TAC1 | 7000195 380706 | 9968 | -0.005 | -0.3934 | No | ||
| 48 | GALR1 | 6020452 | 10191 | -0.006 | -0.4048 | No | ||
| 49 | PTGFR | 3850373 | 10192 | -0.006 | -0.4041 | No | ||
| 50 | P2RY4 | 630053 | 10402 | -0.007 | -0.4147 | No | ||
| 51 | TSHR | 1190687 5270364 | 10721 | -0.008 | -0.4311 | No | ||
| 52 | HCRT | 2970463 | 10755 | -0.008 | -0.4320 | No | ||
| 53 | CCR3 | 50427 | 10780 | -0.008 | -0.4324 | No | ||
| 54 | TACR3 | 4780017 | 10811 | -0.008 | -0.4332 | No | ||
| 55 | HCRTR2 | 2350463 | 10833 | -0.008 | -0.4335 | No | ||
| 56 | CCKBR | 2760128 | 10913 | -0.009 | -0.4369 | No | ||
| 57 | HTR1F | 670148 | 10973 | -0.009 | -0.4391 | No | ||
| 58 | OXT | 3850692 | 11047 | -0.009 | -0.4421 | No | ||
| 59 | CGB | 580273 | 11057 | -0.009 | -0.4417 | No | ||
| 60 | HRH1 | 840100 | 11060 | -0.009 | -0.4408 | No | ||
| 61 | FSHR | 610041 7040121 | 11143 | -0.009 | -0.4443 | No | ||
| 62 | DARC | 3940050 | 11233 | -0.010 | -0.4481 | No | ||
| 63 | NPFF | 2030280 | 11272 | -0.010 | -0.4491 | No | ||
| 64 | AGTR1 | 4780524 2680592 | 11345 | -0.010 | -0.4519 | No | ||
| 65 | CHRM5 | 5390333 | 11514 | -0.011 | -0.4598 | No | ||
| 66 | IL8RA | 1580100 | 11646 | -0.012 | -0.4656 | No | ||
| 67 | PNOC | 1990114 | 11962 | -0.014 | -0.4812 | No | ||
| 68 | CX3CL1 | 3990707 | 12044 | -0.014 | -0.4841 | No | ||
| 69 | ADORA1 | 6370520 | 12105 | -0.015 | -0.4858 | No | ||
| 70 | P2RY1 | 6040121 | 12265 | -0.016 | -0.4927 | No | ||
| 71 | KNG1 | 6400576 6770347 | 12499 | -0.018 | -0.5034 | No | ||
| 72 | AVPR2 | 360064 | 12757 | -0.020 | -0.5152 | No | ||
| 73 | TSHB | 6770500 | 12764 | -0.020 | -0.5134 | No | ||
| 74 | HTR2A | 360037 6330255 | 13104 | -0.024 | -0.5292 | No | ||
| 75 | AGT | 7000575 | 13334 | -0.027 | -0.5388 | No | ||
| 76 | P2RY2 | 2640180 2810128 | 13388 | -0.028 | -0.5387 | No | ||
| 77 | MC5R | 2640176 | 13413 | -0.028 | -0.5371 | No | ||
| 78 | PTGIR | 2100563 | 13831 | -0.036 | -0.5559 | No | ||
| 79 | P2RY14 | 6100497 | 14215 | -0.045 | -0.5719 | No | ||
| 80 | BDKRB1 | 730592 | 15016 | -0.073 | -0.6076 | No | ||
| 81 | ADORA2A | 1990687 | 15332 | -0.090 | -0.6152 | No | ||
| 82 | PDYN | 5720408 | 15587 | -0.112 | -0.6172 | Yes | ||
| 83 | EDNRA | 6900133 | 15620 | -0.114 | -0.6071 | Yes | ||
| 84 | HRH2 | 3840440 | 15833 | -0.135 | -0.6044 | Yes | ||
| 85 | GPR17 | 5890671 | 15926 | -0.146 | -0.5941 | Yes | ||
| 86 | NTSR1 | 6510136 | 15931 | -0.148 | -0.5789 | Yes | ||
| 87 | NMB | 3360735 | 16676 | -0.252 | -0.5928 | Yes | ||
| 88 | F2 | 5720280 | 16781 | -0.270 | -0.5703 | Yes | ||
| 89 | CCR7 | 2060687 | 17376 | -0.388 | -0.5619 | Yes | ||
| 90 | TBXA2R | 2370292 3940066 | 17508 | -0.429 | -0.5242 | Yes | ||
| 91 | CNR2 | 2760398 | 18201 | -0.835 | -0.4746 | Yes | ||
| 92 | ADRB2 | 3290373 | 18208 | -0.843 | -0.3871 | Yes | ||
| 93 | GAL | 3930279 | 18601 | -3.926 | 0.0008 | Yes |