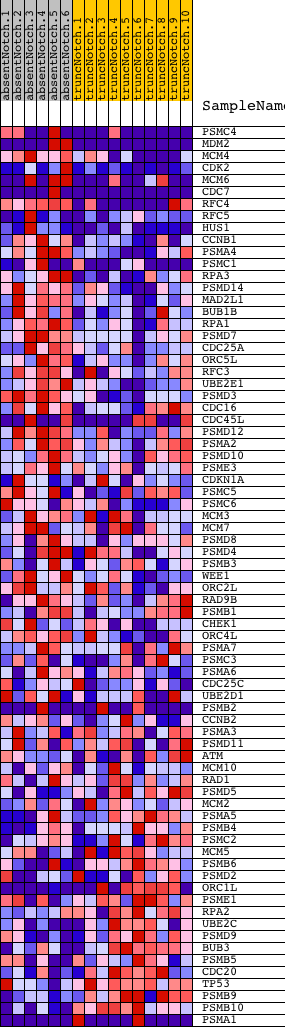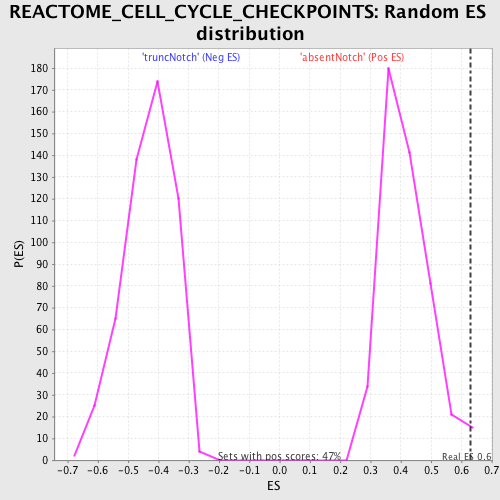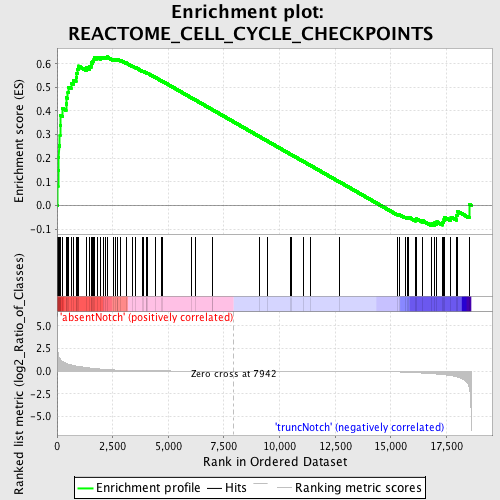
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | REACTOME_CELL_CYCLE_CHECKPOINTS |
| Enrichment Score (ES) | 0.6291397 |
| Normalized Enrichment Score (NES) | 1.5117402 |
| Nominal p-value | 0.010593221 |
| FDR q-value | 0.31936297 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMC4 | 580050 1580025 | 16 | 2.570 | 0.0832 | Yes | ||
| 2 | MDM2 | 3450053 5080138 | 41 | 2.078 | 0.1500 | Yes | ||
| 3 | MCM4 | 2760673 5420711 | 68 | 1.651 | 0.2026 | Yes | ||
| 4 | CDK2 | 130484 2260301 4010088 5050110 | 84 | 1.554 | 0.2526 | Yes | ||
| 5 | MCM6 | 60092 540181 6510110 | 128 | 1.370 | 0.2952 | Yes | ||
| 6 | CDC7 | 4060546 4850041 | 133 | 1.359 | 0.3394 | Yes | ||
| 7 | RFC4 | 3800082 6840142 | 145 | 1.314 | 0.3818 | Yes | ||
| 8 | RFC5 | 3800452 6020091 | 236 | 1.048 | 0.4113 | Yes | ||
| 9 | HUS1 | 3440494 3800156 | 402 | 0.845 | 0.4300 | Yes | ||
| 10 | CCNB1 | 4590433 4780372 | 427 | 0.824 | 0.4557 | Yes | ||
| 11 | PSMA4 | 4560427 | 485 | 0.771 | 0.4779 | Yes | ||
| 12 | PSMC1 | 6350538 | 511 | 0.755 | 0.5012 | Yes | ||
| 13 | RPA3 | 5700136 | 630 | 0.672 | 0.5169 | Yes | ||
| 14 | PSMD14 | 5690593 | 743 | 0.611 | 0.5308 | Yes | ||
| 15 | MAD2L1 | 4480725 | 852 | 0.562 | 0.5434 | Yes | ||
| 16 | BUB1B | 1450288 | 881 | 0.551 | 0.5599 | Yes | ||
| 17 | RPA1 | 360452 | 914 | 0.539 | 0.5758 | Yes | ||
| 18 | PSMD7 | 2030619 6220594 | 956 | 0.515 | 0.5904 | Yes | ||
| 19 | CDC25A | 3800184 | 1320 | 0.382 | 0.5834 | Yes | ||
| 20 | ORC5L | 1940133 1940711 | 1454 | 0.347 | 0.5875 | Yes | ||
| 21 | RFC3 | 1980600 | 1527 | 0.329 | 0.5944 | Yes | ||
| 22 | UBE2E1 | 4230064 1090440 | 1545 | 0.325 | 0.6042 | Yes | ||
| 23 | PSMD3 | 1400647 | 1603 | 0.305 | 0.6111 | Yes | ||
| 24 | CDC16 | 1940706 | 1654 | 0.291 | 0.6179 | Yes | ||
| 25 | CDC45L | 70537 3130114 | 1679 | 0.285 | 0.6259 | Yes | ||
| 26 | PSMD12 | 730044 | 1793 | 0.259 | 0.6283 | Yes | ||
| 27 | PSMA2 | 6510093 | 1957 | 0.220 | 0.6267 | Yes | ||
| 28 | PSMD10 | 520494 1170576 3830050 | 2081 | 0.195 | 0.6265 | Yes | ||
| 29 | PSME3 | 2810537 | 2159 | 0.180 | 0.6282 | Yes | ||
| 30 | CDKN1A | 4050088 6400706 | 2242 | 0.164 | 0.6291 | Yes | ||
| 31 | PSMC5 | 2760315 6550021 | 2521 | 0.125 | 0.6182 | No | ||
| 32 | PSMC6 | 2810021 | 2614 | 0.117 | 0.6171 | No | ||
| 33 | MCM3 | 5570068 | 2691 | 0.107 | 0.6165 | No | ||
| 34 | MCM7 | 3290292 5220056 | 2733 | 0.103 | 0.6177 | No | ||
| 35 | PSMD8 | 630142 | 2853 | 0.093 | 0.6143 | No | ||
| 36 | PSMD4 | 430068 | 3096 | 0.073 | 0.6037 | No | ||
| 37 | PSMB3 | 4280594 | 3387 | 0.055 | 0.5898 | No | ||
| 38 | WEE1 | 3390070 | 3537 | 0.047 | 0.5833 | No | ||
| 39 | ORC2L | 1990470 6510019 | 3821 | 0.035 | 0.5692 | No | ||
| 40 | RAD9B | 2320064 | 3870 | 0.033 | 0.5677 | No | ||
| 41 | PSMB1 | 2940402 | 4007 | 0.028 | 0.5613 | No | ||
| 42 | CHEK1 | 50167 1940300 | 4014 | 0.028 | 0.5619 | No | ||
| 43 | ORC4L | 4230538 5550288 | 4055 | 0.027 | 0.5606 | No | ||
| 44 | PSMA7 | 2230131 | 4405 | 0.019 | 0.5424 | No | ||
| 45 | PSMC3 | 2480184 | 4709 | 0.015 | 0.5266 | No | ||
| 46 | PSMA6 | 50609 | 4732 | 0.014 | 0.5258 | No | ||
| 47 | CDC25C | 2570673 4760161 6520707 | 6018 | 0.006 | 0.4567 | No | ||
| 48 | UBE2D1 | 1850400 | 6220 | 0.005 | 0.4461 | No | ||
| 49 | PSMB2 | 940035 4210324 | 6985 | 0.003 | 0.4049 | No | ||
| 50 | CCNB2 | 6510528 | 6995 | 0.002 | 0.4045 | No | ||
| 51 | PSMA3 | 5900047 7040161 | 9077 | -0.003 | 0.2924 | No | ||
| 52 | PSMD11 | 2340538 6510053 | 9438 | -0.004 | 0.2731 | No | ||
| 53 | ATM | 3610110 4050524 | 10496 | -0.007 | 0.2163 | No | ||
| 54 | MCM10 | 4920632 | 10531 | -0.007 | 0.2147 | No | ||
| 55 | RAD1 | 4200551 | 11076 | -0.009 | 0.1857 | No | ||
| 56 | PSMD5 | 3940139 4570041 | 11393 | -0.011 | 0.1690 | No | ||
| 57 | MCM2 | 5050139 | 12705 | -0.020 | 0.0989 | No | ||
| 58 | PSMA5 | 5390537 | 15296 | -0.088 | -0.0378 | No | ||
| 59 | PSMB4 | 520402 | 15373 | -0.094 | -0.0389 | No | ||
| 60 | PSMC2 | 7040010 | 15638 | -0.116 | -0.0493 | No | ||
| 61 | MCM5 | 2680647 | 15739 | -0.127 | -0.0506 | No | ||
| 62 | PSMB6 | 2690711 | 15804 | -0.133 | -0.0497 | No | ||
| 63 | PSMD2 | 4670706 5050364 | 16110 | -0.172 | -0.0605 | No | ||
| 64 | ORC1L | 2370328 6110390 | 16154 | -0.179 | -0.0569 | No | ||
| 65 | PSME1 | 450193 4480035 | 16418 | -0.217 | -0.0640 | No | ||
| 66 | RPA2 | 2760301 5420195 | 16845 | -0.280 | -0.0778 | No | ||
| 67 | UBE2C | 6130017 | 16960 | -0.302 | -0.0741 | No | ||
| 68 | PSMD9 | 50020 5890368 | 17072 | -0.321 | -0.0696 | No | ||
| 69 | BUB3 | 3170546 | 17321 | -0.375 | -0.0707 | No | ||
| 70 | PSMB5 | 6290242 | 17386 | -0.392 | -0.0613 | No | ||
| 71 | CDC20 | 3440017 3440044 6220088 | 17407 | -0.398 | -0.0494 | No | ||
| 72 | TP53 | 6130707 | 17702 | -0.490 | -0.0492 | No | ||
| 73 | PSMB9 | 6980471 | 17941 | -0.608 | -0.0421 | No | ||
| 74 | PSMB10 | 630504 6510039 | 17986 | -0.631 | -0.0239 | No | ||
| 75 | PSMA1 | 380059 2760195 | 18522 | -1.765 | 0.0051 | No |