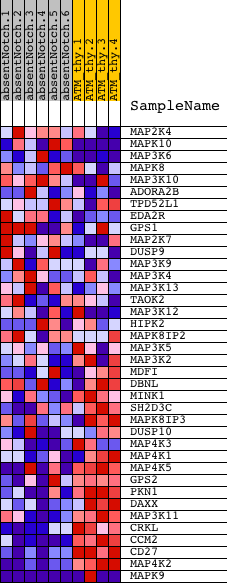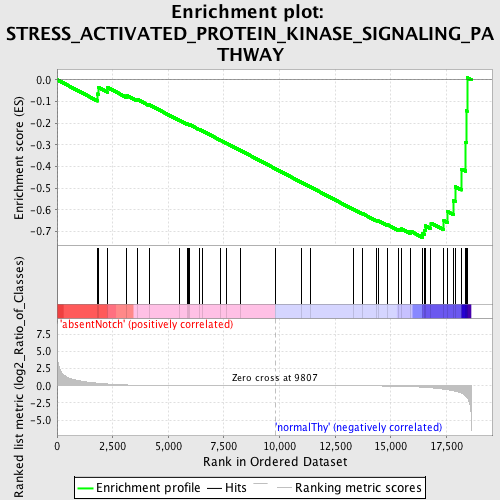
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | STRESS_ACTIVATED_PROTEIN_KINASE_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.72705466 |
| Normalized Enrichment Score (NES) | -1.5129968 |
| Nominal p-value | 0.018518519 |
| FDR q-value | 0.5166981 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP2K4 | 5130133 | 1821 | 0.367 | -0.0655 | No | ||
| 2 | MAPK10 | 6110193 | 1857 | 0.358 | -0.0357 | No | ||
| 3 | MAP3K6 | 6400390 | 2275 | 0.249 | -0.0360 | No | ||
| 4 | MAPK8 | 2640195 | 3135 | 0.114 | -0.0722 | No | ||
| 5 | MAP3K10 | 3360168 4850180 | 3613 | 0.075 | -0.0912 | No | ||
| 6 | ADORA2B | 6860253 | 4144 | 0.048 | -0.1155 | No | ||
| 7 | TPD52L1 | 4590711 5570575 | 5494 | 0.019 | -0.1864 | No | ||
| 8 | EDA2R | 870397 | 5842 | 0.016 | -0.2037 | No | ||
| 9 | GPS1 | 2470037 4760315 6200528 7100040 | 5894 | 0.016 | -0.2050 | No | ||
| 10 | MAP2K7 | 2260086 | 5971 | 0.015 | -0.2078 | No | ||
| 11 | DUSP9 | 630605 | 6400 | 0.012 | -0.2298 | No | ||
| 12 | MAP3K9 | 3290707 | 6538 | 0.011 | -0.2361 | No | ||
| 13 | MAP3K4 | 5720041 | 7343 | 0.008 | -0.2787 | No | ||
| 14 | MAP3K13 | 3190017 | 7593 | 0.007 | -0.2916 | No | ||
| 15 | TAOK2 | 110575 6380692 | 8258 | 0.004 | -0.3269 | No | ||
| 16 | MAP3K12 | 2470373 6420162 | 9831 | -0.000 | -0.4115 | No | ||
| 17 | HIPK2 | 6350647 | 10981 | -0.003 | -0.4730 | No | ||
| 18 | MAPK8IP2 | 630373 | 11370 | -0.005 | -0.4935 | No | ||
| 19 | MAP3K5 | 6020041 6380162 | 13343 | -0.015 | -0.5983 | No | ||
| 20 | MAP3K2 | 380270 | 13729 | -0.019 | -0.6174 | No | ||
| 21 | MDFI | 6200041 | 14358 | -0.029 | -0.6486 | No | ||
| 22 | DBNL | 7100088 | 14452 | -0.031 | -0.6509 | No | ||
| 23 | MINK1 | 5860647 | 14867 | -0.045 | -0.6692 | No | ||
| 24 | SH2D3C | 3060014 | 15344 | -0.075 | -0.6881 | No | ||
| 25 | MAPK8IP3 | 1660112 2060427 | 15477 | -0.087 | -0.6875 | No | ||
| 26 | DUSP10 | 2850673 3360064 | 15882 | -0.129 | -0.6978 | No | ||
| 27 | MAP4K3 | 5390551 | 16426 | -0.209 | -0.7085 | Yes | ||
| 28 | MAP4K1 | 4060692 | 16525 | -0.231 | -0.6933 | Yes | ||
| 29 | MAP4K5 | 3190286 4760300 5270070 5550494 | 16556 | -0.239 | -0.6738 | Yes | ||
| 30 | GPS2 | 5910672 | 16804 | -0.295 | -0.6609 | Yes | ||
| 31 | PKN1 | 2640605 2760647 | 17382 | -0.487 | -0.6488 | Yes | ||
| 32 | DAXX | 4010647 | 17546 | -0.560 | -0.6080 | Yes | ||
| 33 | MAP3K11 | 7000039 | 17815 | -0.725 | -0.5582 | Yes | ||
| 34 | CRKL | 4050427 | 17897 | -0.783 | -0.4932 | Yes | ||
| 35 | CCM2 | 5670465 | 18178 | -1.092 | -0.4116 | Yes | ||
| 36 | CD27 | 610020 | 18360 | -1.500 | -0.2885 | Yes | ||
| 37 | MAP4K2 | 4200037 | 18402 | -1.668 | -0.1430 | Yes | ||
| 38 | MAPK9 | 2060273 3780209 4070397 | 18430 | -1.744 | 0.0100 | Yes |