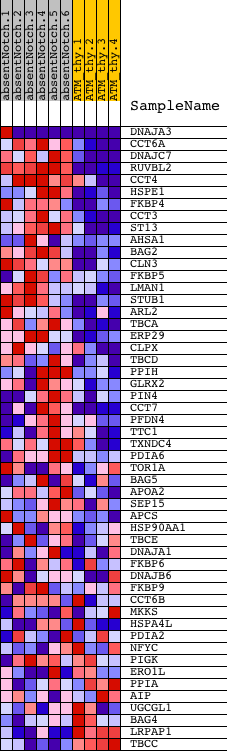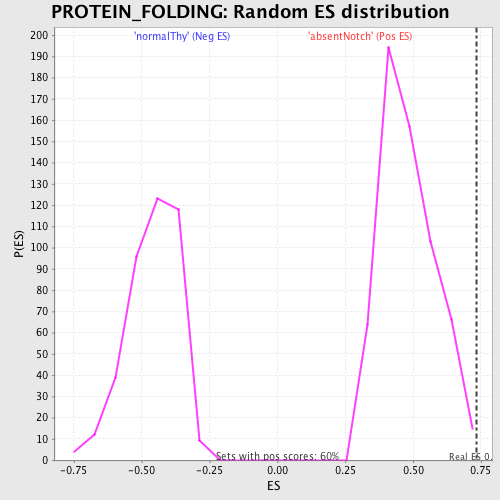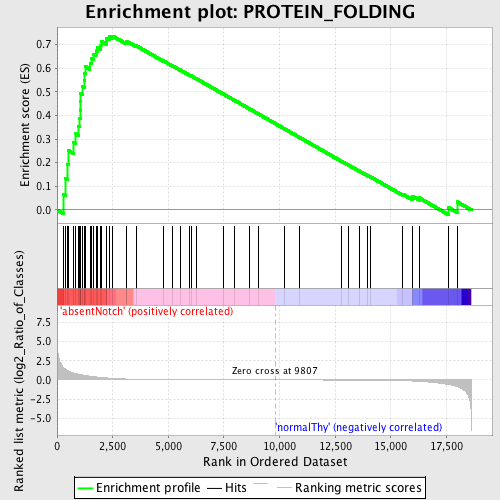
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | PROTEIN_FOLDING |
| Enrichment Score (ES) | 0.735425 |
| Normalized Enrichment Score (NES) | 1.5336436 |
| Nominal p-value | 0.0050083473 |
| FDR q-value | 0.27193633 |
| FWER p-Value | 0.946 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DNAJA3 | 1980671 6220338 | 289 | 1.568 | 0.0661 | Yes | ||
| 2 | CCT6A | 2350368 | 366 | 1.398 | 0.1349 | Yes | ||
| 3 | DNAJC7 | 4920048 | 465 | 1.225 | 0.1935 | Yes | ||
| 4 | RUVBL2 | 1190377 | 504 | 1.152 | 0.2515 | Yes | ||
| 5 | CCT4 | 6380161 | 752 | 0.883 | 0.2842 | Yes | ||
| 6 | HSPE1 | 3170438 | 845 | 0.815 | 0.3217 | Yes | ||
| 7 | FKBP4 | 870035 | 954 | 0.748 | 0.3548 | Yes | ||
| 8 | CCT3 | 7100403 | 1021 | 0.701 | 0.3878 | Yes | ||
| 9 | ST13 | 3850369 | 1037 | 0.691 | 0.4230 | Yes | ||
| 10 | AHSA1 | 2030520 | 1045 | 0.688 | 0.4585 | Yes | ||
| 11 | BAG2 | 2900324 | 1051 | 0.685 | 0.4939 | Yes | ||
| 12 | CLN3 | 780368 | 1147 | 0.635 | 0.5219 | Yes | ||
| 13 | FKBP5 | 2190048 | 1218 | 0.596 | 0.5491 | Yes | ||
| 14 | LMAN1 | 3420068 | 1238 | 0.587 | 0.5787 | Yes | ||
| 15 | STUB1 | 5860086 | 1275 | 0.569 | 0.6064 | Yes | ||
| 16 | ARL2 | 5890377 | 1478 | 0.484 | 0.6207 | Yes | ||
| 17 | TBCA | 6400095 | 1537 | 0.460 | 0.6416 | Yes | ||
| 18 | ERP29 | 4050441 | 1638 | 0.425 | 0.6584 | Yes | ||
| 19 | CLPX | 6220156 | 1753 | 0.393 | 0.6727 | Yes | ||
| 20 | TBCD | 5720020 | 1822 | 0.367 | 0.6881 | Yes | ||
| 21 | PPIH | 5910403 | 1966 | 0.326 | 0.6974 | Yes | ||
| 22 | GLRX2 | 940433 | 1981 | 0.323 | 0.7135 | Yes | ||
| 23 | PIN4 | 2370324 | 2226 | 0.258 | 0.7138 | Yes | ||
| 24 | CCT7 | 6660091 | 2232 | 0.257 | 0.7269 | Yes | ||
| 25 | PFDN4 | 4010286 | 2348 | 0.232 | 0.7328 | Yes | ||
| 26 | TTC1 | 5890736 | 2497 | 0.203 | 0.7354 | Yes | ||
| 27 | TXNDC4 | 450537 | 3105 | 0.116 | 0.7088 | No | ||
| 28 | PDIA6 | 2760632 | 3131 | 0.114 | 0.7134 | No | ||
| 29 | TOR1A | 670139 5890288 | 3552 | 0.079 | 0.6949 | No | ||
| 30 | BAG5 | 2230368 6420537 | 4766 | 0.030 | 0.6311 | No | ||
| 31 | APOA2 | 6510364 6860411 | 5202 | 0.023 | 0.6088 | No | ||
| 32 | SEP15 | 1780451 2060450 5890451 | 5527 | 0.019 | 0.5923 | No | ||
| 33 | APCS | 2060500 | 5950 | 0.015 | 0.5704 | No | ||
| 34 | HSP90AA1 | 4560041 5220133 2120722 | 6024 | 0.015 | 0.5672 | No | ||
| 35 | TBCE | 520692 | 6266 | 0.013 | 0.5549 | No | ||
| 36 | DNAJA1 | 1660010 2320358 6840136 | 7468 | 0.007 | 0.4906 | No | ||
| 37 | FKBP6 | 6840725 | 7959 | 0.005 | 0.4645 | No | ||
| 38 | DNAJB6 | 2810309 5570451 | 8625 | 0.003 | 0.4288 | No | ||
| 39 | FKBP9 | 1400044 | 9071 | 0.002 | 0.4050 | No | ||
| 40 | CCT6B | 3190411 | 10210 | -0.001 | 0.3437 | No | ||
| 41 | MKKS | 2760722 3390563 | 10891 | -0.003 | 0.3073 | No | ||
| 42 | HSPA4L | 5550035 | 12773 | -0.011 | 0.2065 | No | ||
| 43 | PDIA2 | 5360706 4670300 5550040 | 13084 | -0.013 | 0.1905 | No | ||
| 44 | NFYC | 1400746 | 13572 | -0.017 | 0.1652 | No | ||
| 45 | PIGK | 4540193 6350324 6860736 | 13973 | -0.022 | 0.1448 | No | ||
| 46 | ERO1L | 3060242 4780154 5220075 | 14065 | -0.024 | 0.1411 | No | ||
| 47 | PPIA | 2900368 4850162 | 15543 | -0.094 | 0.0665 | No | ||
| 48 | AIP | 5720300 | 15966 | -0.139 | 0.0510 | No | ||
| 49 | UGCGL1 | 4230072 | 15980 | -0.141 | 0.0576 | No | ||
| 50 | BAG4 | 2260070 | 16270 | -0.180 | 0.0514 | No | ||
| 51 | LRPAP1 | 5570253 | 17579 | -0.580 | 0.0112 | No | ||
| 52 | TBCC | 4810021 | 17983 | -0.856 | 0.0341 | No |