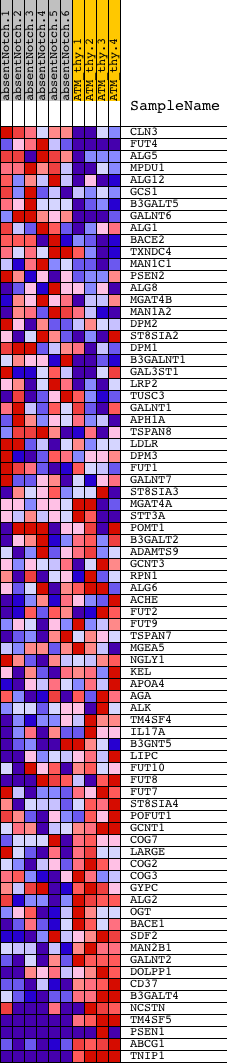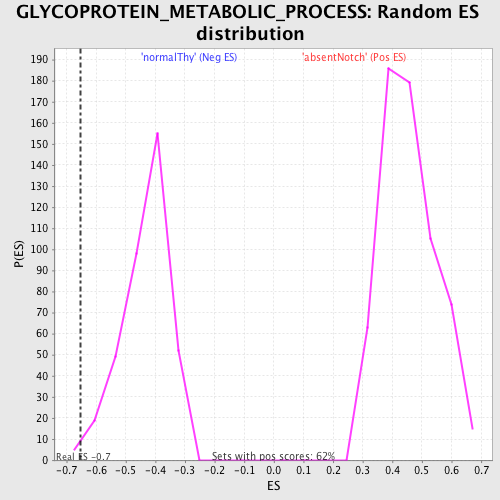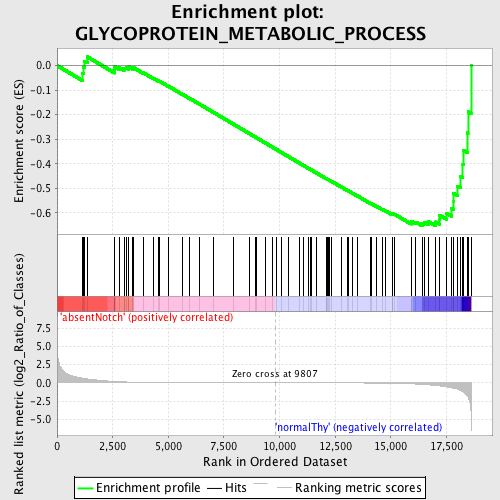
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GLYCOPROTEIN_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.65328586 |
| Normalized Enrichment Score (NES) | -1.5119463 |
| Nominal p-value | 0.007936508 |
| FDR q-value | 0.47792548 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CLN3 | 780368 | 1147 | 0.635 | -0.0328 | No | ||
| 2 | FUT4 | 2030324 | 1175 | 0.618 | -0.0060 | No | ||
| 3 | ALG5 | 3060692 4780093 6590097 | 1228 | 0.592 | 0.0183 | No | ||
| 4 | MPDU1 | 1850091 | 1361 | 0.527 | 0.0353 | No | ||
| 5 | ALG12 | 1400692 | 2581 | 0.189 | -0.0218 | No | ||
| 6 | GCS1 | 1740603 | 2591 | 0.187 | -0.0137 | No | ||
| 7 | B3GALT5 | 4060047 | 2599 | 0.185 | -0.0056 | No | ||
| 8 | GALNT6 | 3710377 | 2793 | 0.155 | -0.0090 | No | ||
| 9 | ALG1 | 2570112 | 3010 | 0.127 | -0.0148 | No | ||
| 10 | BACE2 | 4060170 5690100 6220133 | 3031 | 0.125 | -0.0102 | No | ||
| 11 | TXNDC4 | 450537 | 3105 | 0.116 | -0.0088 | No | ||
| 12 | MAN1C1 | 730133 870398 2340044 | 3220 | 0.105 | -0.0101 | No | ||
| 13 | PSEN2 | 130382 | 3230 | 0.104 | -0.0058 | No | ||
| 14 | ALG8 | 4150497 4780500 6290563 | 3395 | 0.090 | -0.0105 | No | ||
| 15 | MGAT4B | 130053 6840100 | 3430 | 0.087 | -0.0084 | No | ||
| 16 | MAN1A2 | 5700435 5720114 | 3882 | 0.059 | -0.0300 | No | ||
| 17 | DPM2 | 3610253 | 4323 | 0.042 | -0.0518 | No | ||
| 18 | ST8SIA2 | 2680082 | 4545 | 0.035 | -0.0621 | No | ||
| 19 | DPM1 | 1240050 4810019 6380398 | 4594 | 0.034 | -0.0632 | No | ||
| 20 | B3GALNT1 | 3390746 60131 | 4990 | 0.026 | -0.0833 | No | ||
| 21 | GAL3ST1 | 5080746 | 5642 | 0.018 | -0.1176 | No | ||
| 22 | LRP2 | 1400204 3290706 | 5929 | 0.015 | -0.1323 | No | ||
| 23 | TUSC3 | 1690411 | 6414 | 0.012 | -0.1579 | No | ||
| 24 | GALNT1 | 4070487 6130100 6250132 6900048 | 7010 | 0.009 | -0.1896 | No | ||
| 25 | APH1A | 4060273 4670465 6450711 | 7934 | 0.005 | -0.2391 | No | ||
| 26 | TSPAN8 | 1740538 | 8631 | 0.003 | -0.2765 | No | ||
| 27 | LDLR | 5670386 | 8928 | 0.002 | -0.2923 | No | ||
| 28 | DPM3 | 50039 | 8976 | 0.002 | -0.2948 | No | ||
| 29 | FUT1 | 1500068 | 9350 | 0.001 | -0.3148 | No | ||
| 30 | GALNT7 | 3990280 | 9669 | 0.000 | -0.3320 | No | ||
| 31 | ST8SIA3 | 6940427 | 9878 | -0.000 | -0.3432 | No | ||
| 32 | MGAT4A | 5690471 | 10069 | -0.001 | -0.3534 | No | ||
| 33 | STT3A | 580139 | 10410 | -0.002 | -0.3716 | No | ||
| 34 | POMT1 | 2680047 3440040 6660114 | 10915 | -0.003 | -0.3987 | No | ||
| 35 | B3GALT2 | 430136 | 11078 | -0.004 | -0.4072 | No | ||
| 36 | ADAMTS9 | 2760079 | 11308 | -0.005 | -0.4194 | No | ||
| 37 | GCNT3 | 6290239 | 11400 | -0.005 | -0.4241 | No | ||
| 38 | RPN1 | 1240113 | 11442 | -0.005 | -0.4261 | No | ||
| 39 | ALG6 | 4060131 | 11667 | -0.006 | -0.4379 | No | ||
| 40 | ACHE | 5290750 | 12092 | -0.008 | -0.4604 | No | ||
| 41 | FUT2 | 3990047 | 12109 | -0.008 | -0.4609 | No | ||
| 42 | FUT9 | 3940121 | 12163 | -0.008 | -0.4634 | No | ||
| 43 | TSPAN7 | 870133 | 12203 | -0.008 | -0.4651 | No | ||
| 44 | MGEA5 | 6100014 | 12244 | -0.008 | -0.4669 | No | ||
| 45 | NGLY1 | 1570037 | 12354 | -0.009 | -0.4724 | No | ||
| 46 | KEL | 6940440 | 12775 | -0.011 | -0.4946 | No | ||
| 47 | APOA4 | 4120451 | 13061 | -0.013 | -0.5093 | No | ||
| 48 | AGA | 6350292 | 13080 | -0.013 | -0.5097 | No | ||
| 49 | ALK | 730008 | 13284 | -0.014 | -0.5200 | No | ||
| 50 | TM4SF4 | 540600 | 13520 | -0.017 | -0.5319 | No | ||
| 51 | IL17A | 4280369 | 14067 | -0.024 | -0.5603 | No | ||
| 52 | B3GNT5 | 4780528 | 14137 | -0.025 | -0.5629 | No | ||
| 53 | LIPC | 2690497 | 14343 | -0.029 | -0.5726 | No | ||
| 54 | FUT10 | 6510603 | 14634 | -0.036 | -0.5866 | No | ||
| 55 | FUT8 | 1340068 2340056 | 14780 | -0.041 | -0.5925 | No | ||
| 56 | FUT7 | 2900736 | 15054 | -0.055 | -0.6047 | No | ||
| 57 | ST8SIA4 | 2680605 3060215 | 15082 | -0.057 | -0.6036 | No | ||
| 58 | POFUT1 | 1570458 | 15165 | -0.062 | -0.6052 | No | ||
| 59 | GCNT1 | 940129 1780215 2370253 | 15910 | -0.132 | -0.6393 | No | ||
| 60 | COG7 | 360133 | 15920 | -0.132 | -0.6337 | No | ||
| 61 | LARGE | 6940068 | 16123 | -0.161 | -0.6373 | No | ||
| 62 | COG2 | 5900129 | 16421 | -0.209 | -0.6437 | Yes | ||
| 63 | COG3 | 3840746 | 16493 | -0.222 | -0.6374 | Yes | ||
| 64 | GYPC | 2320102 | 16686 | -0.267 | -0.6355 | Yes | ||
| 65 | ALG2 | 3990091 | 17013 | -0.351 | -0.6370 | Yes | ||
| 66 | OGT | 2360131 4610333 | 17176 | -0.406 | -0.6272 | Yes | ||
| 67 | BACE1 | 2650438 | 17187 | -0.409 | -0.6090 | Yes | ||
| 68 | SDF2 | 2690601 | 17496 | -0.536 | -0.6010 | Yes | ||
| 69 | MAN2B1 | 4210446 | 17722 | -0.670 | -0.5825 | Yes | ||
| 70 | GALNT2 | 2260279 4060239 | 17819 | -0.727 | -0.5544 | Yes | ||
| 71 | DOLPP1 | 4780619 6400053 | 17823 | -0.728 | -0.5213 | Yes | ||
| 72 | CD37 | 770273 | 17996 | -0.869 | -0.4908 | Yes | ||
| 73 | B3GALT4 | 2340239 | 18126 | -1.020 | -0.4510 | Yes | ||
| 74 | NCSTN | 940601 6400594 | 18240 | -1.190 | -0.4027 | Yes | ||
| 75 | TM4SF5 | 3120280 | 18277 | -1.287 | -0.3457 | Yes | ||
| 76 | PSEN1 | 130403 2030647 6100603 | 18435 | -1.758 | -0.2737 | Yes | ||
| 77 | ABCG1 | 60692 | 18470 | -1.918 | -0.1877 | Yes | ||
| 78 | TNIP1 | 2510279 5900041 6510435 | 18608 | -4.273 | 0.0004 | Yes |