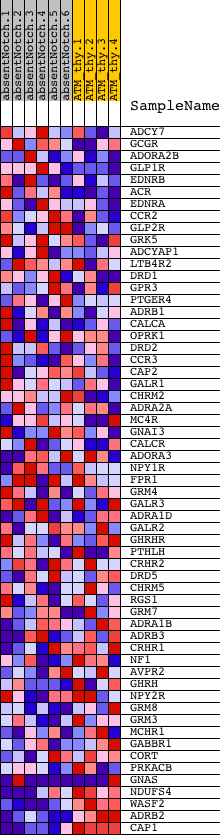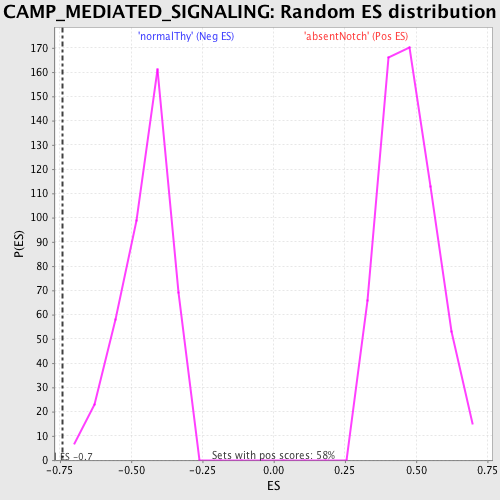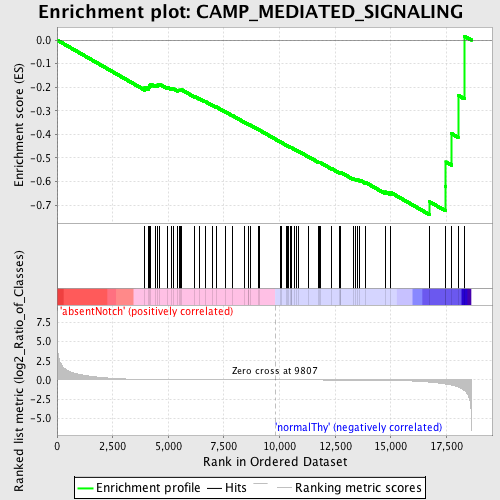
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CAMP_MEDIATED_SIGNALING |
| Enrichment Score (ES) | -0.7397917 |
| Normalized Enrichment Score (NES) | -1.6454154 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.196273 |
| FWER p-Value | 0.346 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADCY7 | 6290520 7560739 | 3945 | 0.056 | -0.2014 | No | ||
| 2 | GCGR | 6620497 | 4116 | 0.049 | -0.2009 | No | ||
| 3 | ADORA2B | 6860253 | 4144 | 0.048 | -0.1928 | No | ||
| 4 | GLP1R | 6420528 | 4210 | 0.046 | -0.1873 | No | ||
| 5 | EDNRB | 4280717 6400435 | 4421 | 0.038 | -0.1910 | No | ||
| 6 | ACR | 2940017 3060050 | 4533 | 0.036 | -0.1899 | No | ||
| 7 | EDNRA | 6900133 | 4616 | 0.033 | -0.1877 | No | ||
| 8 | CCR2 | 1450519 | 4940 | 0.027 | -0.1998 | No | ||
| 9 | GLP2R | 4150577 5290273 7050095 | 5123 | 0.024 | -0.2049 | No | ||
| 10 | GRK5 | 1940348 4670053 | 5251 | 0.022 | -0.2073 | No | ||
| 11 | ADCYAP1 | 520086 | 5423 | 0.020 | -0.2126 | No | ||
| 12 | LTB4R2 | 1990746 | 5479 | 0.019 | -0.2118 | No | ||
| 13 | DRD1 | 430025 | 5550 | 0.019 | -0.2119 | No | ||
| 14 | GPR3 | 2970452 | 5609 | 0.018 | -0.2114 | No | ||
| 15 | PTGER4 | 6400670 | 6193 | 0.013 | -0.2402 | No | ||
| 16 | ADRB1 | 6860292 | 6408 | 0.012 | -0.2493 | No | ||
| 17 | CALCA | 5860167 | 6655 | 0.011 | -0.2604 | No | ||
| 18 | OPRK1 | 3780632 | 6974 | 0.009 | -0.2758 | No | ||
| 19 | DRD2 | 5890369 | 7145 | 0.008 | -0.2833 | No | ||
| 20 | CCR3 | 50427 | 7564 | 0.007 | -0.3045 | No | ||
| 21 | CAP2 | 7100315 | 7892 | 0.006 | -0.3210 | No | ||
| 22 | GALR1 | 6020452 | 8429 | 0.004 | -0.3491 | No | ||
| 23 | CHRM2 | 870750 | 8609 | 0.003 | -0.3580 | No | ||
| 24 | ADRA2A | 5340520 | 8620 | 0.003 | -0.3579 | No | ||
| 25 | MC4R | 430333 | 8675 | 0.003 | -0.3602 | No | ||
| 26 | GNAI3 | 4670739 | 9052 | 0.002 | -0.3800 | No | ||
| 27 | CALCR | 1690494 | 9114 | 0.002 | -0.3829 | No | ||
| 28 | ADORA3 | 630333 | 10029 | -0.001 | -0.4320 | No | ||
| 29 | NPY1R | 5890347 | 10102 | -0.001 | -0.4357 | No | ||
| 30 | FPR1 | 3800577 | 10332 | -0.002 | -0.4478 | No | ||
| 31 | GRM4 | 6960092 | 10370 | -0.002 | -0.4494 | No | ||
| 32 | GALR3 | 4850113 | 10405 | -0.002 | -0.4509 | No | ||
| 33 | ADRA1D | 380035 540025 | 10479 | -0.002 | -0.4545 | No | ||
| 34 | GALR2 | 6450739 | 10523 | -0.002 | -0.4564 | No | ||
| 35 | GHRHR | 3130500 4010154 | 10543 | -0.002 | -0.4570 | No | ||
| 36 | PTHLH | 5290739 | 10686 | -0.002 | -0.4641 | No | ||
| 37 | CRHR2 | 4590672 | 10739 | -0.003 | -0.4664 | No | ||
| 38 | DRD5 | 2970133 | 10844 | -0.003 | -0.4714 | No | ||
| 39 | CHRM5 | 5390333 | 11304 | -0.005 | -0.4952 | No | ||
| 40 | RGS1 | 4060347 4540181 | 11770 | -0.006 | -0.5191 | No | ||
| 41 | GRM7 | 4230110 | 11793 | -0.006 | -0.5190 | No | ||
| 42 | ADRA1B | 2100484 | 11822 | -0.006 | -0.5193 | No | ||
| 43 | ADRB3 | 6900072 | 12345 | -0.009 | -0.5457 | No | ||
| 44 | CRHR1 | 450161 3140070 5390647 | 12692 | -0.010 | -0.5622 | No | ||
| 45 | NF1 | 6980433 | 12719 | -0.011 | -0.5615 | No | ||
| 46 | AVPR2 | 360064 | 12746 | -0.011 | -0.5608 | No | ||
| 47 | GHRH | 4570575 | 13322 | -0.015 | -0.5888 | No | ||
| 48 | NPY2R | 6220612 | 13410 | -0.015 | -0.5905 | No | ||
| 49 | GRM8 | 4780082 | 13492 | -0.016 | -0.5916 | No | ||
| 50 | GRM3 | 4540242 | 13611 | -0.018 | -0.5944 | No | ||
| 51 | MCHR1 | 1780022 | 13867 | -0.021 | -0.6040 | No | ||
| 52 | GABBR1 | 2850519 2850735 | 14763 | -0.040 | -0.6442 | No | ||
| 53 | CORT | 360008 2340092 | 14981 | -0.051 | -0.6458 | No | ||
| 54 | PRKACB | 4210170 | 16727 | -0.276 | -0.6851 | Yes | ||
| 55 | GNAS | 630441 1850373 4050152 | 17449 | -0.517 | -0.6213 | Yes | ||
| 56 | NDUFS4 | 510142 1240148 4760253 | 17479 | -0.533 | -0.5171 | Yes | ||
| 57 | WASF2 | 4850592 | 17735 | -0.674 | -0.3971 | Yes | ||
| 58 | ADRB2 | 3290373 | 18026 | -0.899 | -0.2344 | Yes | ||
| 59 | CAP1 | 2650278 | 18305 | -1.341 | 0.0168 | Yes |