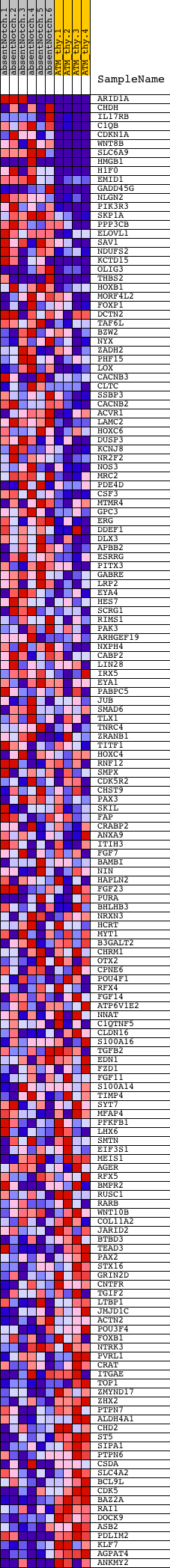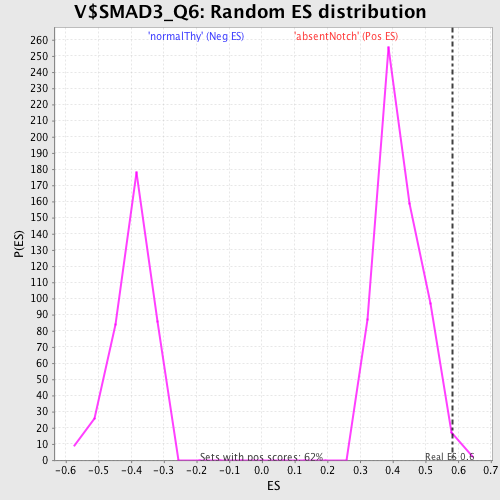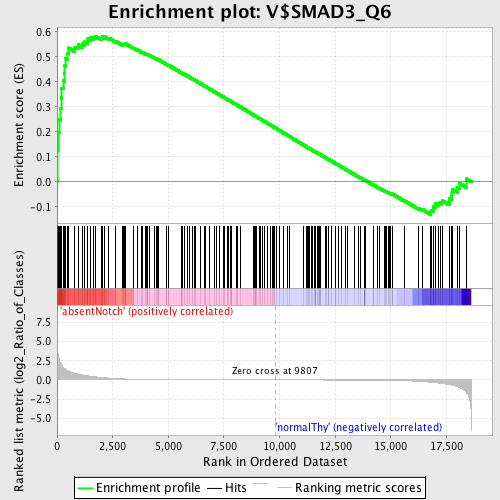
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$SMAD3_Q6 |
| Enrichment Score (ES) | 0.58305013 |
| Normalized Enrichment Score (NES) | 1.3838614 |
| Nominal p-value | 0.009724474 |
| FDR q-value | 0.8724475 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARID1A | 2630022 1690551 4810110 | 7 | 5.913 | 0.1275 | Yes | ||
| 2 | CHDH | 2570711 6200338 | 40 | 3.397 | 0.1992 | Yes | ||
| 3 | IL17RB | 6590215 | 109 | 2.502 | 0.2496 | Yes | ||
| 4 | C1QB | 5910292 | 159 | 2.145 | 0.2933 | Yes | ||
| 5 | CDKN1A | 4050088 6400706 | 188 | 2.027 | 0.3357 | Yes | ||
| 6 | WNT8B | 4560301 | 209 | 1.875 | 0.3751 | Yes | ||
| 7 | SLC6A9 | 2470520 | 276 | 1.588 | 0.4059 | Yes | ||
| 8 | HMGB1 | 2120670 2350044 | 338 | 1.462 | 0.4342 | Yes | ||
| 9 | H1F0 | 1400408 | 348 | 1.437 | 0.4648 | Yes | ||
| 10 | EMID1 | 3190079 5130152 | 385 | 1.364 | 0.4923 | Yes | ||
| 11 | GADD45G | 2510142 | 458 | 1.243 | 0.5153 | Yes | ||
| 12 | NLGN2 | 5900592 | 530 | 1.115 | 0.5356 | Yes | ||
| 13 | PIK3R3 | 1770333 | 800 | 0.839 | 0.5391 | Yes | ||
| 14 | SKP1A | 2450102 | 939 | 0.756 | 0.5480 | Yes | ||
| 15 | PPP3CB | 6020156 | 1128 | 0.643 | 0.5517 | Yes | ||
| 16 | ELOVL1 | 2450019 4760138 5340685 | 1220 | 0.595 | 0.5596 | Yes | ||
| 17 | SAV1 | 5420541 | 1372 | 0.523 | 0.5628 | Yes | ||
| 18 | NDUFS2 | 4850020 6200402 | 1386 | 0.519 | 0.5733 | Yes | ||
| 19 | KCTD15 | 2810735 | 1516 | 0.469 | 0.5765 | Yes | ||
| 20 | OLIG3 | 1050603 | 1654 | 0.419 | 0.5781 | Yes | ||
| 21 | THBS2 | 2850136 | 1723 | 0.400 | 0.5831 | Yes | ||
| 22 | HOXB1 | 2120110 | 1999 | 0.318 | 0.5750 | No | ||
| 23 | MORF4L2 | 6450133 | 2024 | 0.312 | 0.5805 | No | ||
| 24 | FOXP1 | 6510433 | 2131 | 0.282 | 0.5808 | No | ||
| 25 | DCTN2 | 540471 3780717 | 2329 | 0.237 | 0.5753 | No | ||
| 26 | TAF6L | 1850056 | 2630 | 0.179 | 0.5629 | No | ||
| 27 | BZW2 | 940079 | 2925 | 0.136 | 0.5499 | No | ||
| 28 | NYX | 4120239 | 2975 | 0.131 | 0.5501 | No | ||
| 29 | ZADH2 | 520438 | 3011 | 0.127 | 0.5509 | No | ||
| 30 | PHF15 | 6550066 6860673 | 3065 | 0.122 | 0.5507 | No | ||
| 31 | LOX | 5340372 | 3074 | 0.121 | 0.5529 | No | ||
| 32 | CACNB3 | 2480035 4540184 | 3445 | 0.086 | 0.5347 | No | ||
| 33 | CLTC | 6590458 | 3605 | 0.076 | 0.5277 | No | ||
| 34 | SSBP3 | 870717 1050377 7040551 | 3782 | 0.065 | 0.5196 | No | ||
| 35 | CACNB2 | 1500095 7330707 | 3848 | 0.061 | 0.5174 | No | ||
| 36 | ACVR1 | 6840671 | 3959 | 0.056 | 0.5126 | No | ||
| 37 | LAMC2 | 450692 2680041 4200059 4670148 | 4016 | 0.053 | 0.5107 | No | ||
| 38 | HOXC6 | 3850133 4920736 | 4060 | 0.051 | 0.5095 | No | ||
| 39 | DUSP3 | 1240129 6840215 | 4082 | 0.050 | 0.5094 | No | ||
| 40 | KCNJ8 | 870494 | 4143 | 0.048 | 0.5072 | No | ||
| 41 | NR2F2 | 3170609 3310577 | 4398 | 0.040 | 0.4943 | No | ||
| 42 | NOS3 | 630152 670465 | 4451 | 0.038 | 0.4923 | No | ||
| 43 | MRC2 | 110369 2480520 | 4505 | 0.036 | 0.4902 | No | ||
| 44 | PDE4D | 2470528 6660014 | 4558 | 0.035 | 0.4882 | No | ||
| 45 | CSF3 | 2230193 6660707 | 4566 | 0.035 | 0.4885 | No | ||
| 46 | MTMR4 | 4540524 | 4916 | 0.027 | 0.4702 | No | ||
| 47 | GPC3 | 6180735 | 4927 | 0.027 | 0.4703 | No | ||
| 48 | ERG | 50154 1770739 | 5000 | 0.026 | 0.4669 | No | ||
| 49 | DDEF1 | 1170411 4070465 | 5020 | 0.025 | 0.4664 | No | ||
| 50 | DLX3 | 3190497 | 5591 | 0.018 | 0.4359 | No | ||
| 51 | APBB2 | 5130403 | 5604 | 0.018 | 0.4357 | No | ||
| 52 | ESRRG | 4010181 | 5656 | 0.018 | 0.4333 | No | ||
| 53 | PITX3 | 3130162 | 5703 | 0.017 | 0.4312 | No | ||
| 54 | GABRE | 4780020 | 5852 | 0.016 | 0.4235 | No | ||
| 55 | LRP2 | 1400204 3290706 | 5929 | 0.015 | 0.4197 | No | ||
| 56 | EYA4 | 5360286 | 6098 | 0.014 | 0.4109 | No | ||
| 57 | HES7 | 4610239 | 6181 | 0.013 | 0.4067 | No | ||
| 58 | SCRG1 | 2060541 | 6226 | 0.013 | 0.4046 | No | ||
| 59 | RIMS1 | 5670022 | 6451 | 0.012 | 0.3928 | No | ||
| 60 | PAK3 | 4210136 | 6603 | 0.011 | 0.3848 | No | ||
| 61 | ARHGEF19 | 940102 | 6656 | 0.011 | 0.3822 | No | ||
| 62 | NXPH4 | 3830500 | 6857 | 0.010 | 0.3716 | No | ||
| 63 | CABP2 | 840142 | 7063 | 0.009 | 0.3607 | No | ||
| 64 | LIN28 | 6590672 | 7157 | 0.008 | 0.3558 | No | ||
| 65 | IRX5 | 2630494 | 7310 | 0.008 | 0.3477 | No | ||
| 66 | EYA1 | 1450278 5220390 | 7489 | 0.007 | 0.3382 | No | ||
| 67 | PABPC5 | 2320408 | 7542 | 0.007 | 0.3356 | No | ||
| 68 | JUB | 5860711 5900500 | 7669 | 0.006 | 0.3289 | No | ||
| 69 | SMAD6 | 870504 | 7715 | 0.006 | 0.3266 | No | ||
| 70 | TLX1 | 2630162 | 7776 | 0.006 | 0.3235 | No | ||
| 71 | TNRC4 | 4050156 | 7834 | 0.006 | 0.3205 | No | ||
| 72 | ZRANB1 | 510373 4540278 | 7838 | 0.006 | 0.3205 | No | ||
| 73 | TITF1 | 3390554 4540292 | 8062 | 0.005 | 0.3085 | No | ||
| 74 | HOXC4 | 1940193 | 8098 | 0.005 | 0.3067 | No | ||
| 75 | RNF12 | 6860717 | 8227 | 0.005 | 0.2999 | No | ||
| 76 | SMPX | 6590440 | 8821 | 0.003 | 0.2678 | No | ||
| 77 | CDK5R2 | 3800110 | 8851 | 0.003 | 0.2663 | No | ||
| 78 | CHST9 | 4560092 | 8913 | 0.003 | 0.2630 | No | ||
| 79 | PAX3 | 50551 | 8925 | 0.002 | 0.2625 | No | ||
| 80 | SKIL | 1090364 | 8954 | 0.002 | 0.2610 | No | ||
| 81 | FAP | 4560019 | 9115 | 0.002 | 0.2524 | No | ||
| 82 | CRABP2 | 6940725 | 9162 | 0.002 | 0.2499 | No | ||
| 83 | ANXA9 | 1190195 | 9249 | 0.002 | 0.2453 | No | ||
| 84 | ITIH3 | 3830551 | 9328 | 0.001 | 0.2411 | No | ||
| 85 | FGF7 | 5390484 | 9464 | 0.001 | 0.2338 | No | ||
| 86 | BAMBI | 6650463 | 9589 | 0.001 | 0.2271 | No | ||
| 87 | NIN | 3450576 5080048 | 9686 | 0.000 | 0.2219 | No | ||
| 88 | HAPLN2 | 2900647 | 9713 | 0.000 | 0.2205 | No | ||
| 89 | FGF23 | 510746 | 9792 | 0.000 | 0.2163 | No | ||
| 90 | PURA | 3870156 | 9881 | -0.000 | 0.2115 | No | ||
| 91 | BHLHB3 | 3450438 | 10001 | -0.001 | 0.2051 | No | ||
| 92 | NRXN3 | 1400546 3360471 | 10155 | -0.001 | 0.1968 | No | ||
| 93 | HCRT | 2970463 | 10360 | -0.002 | 0.1858 | No | ||
| 94 | MYT1 | 6770524 | 10449 | -0.002 | 0.1811 | No | ||
| 95 | B3GALT2 | 430136 | 11078 | -0.004 | 0.1471 | No | ||
| 96 | CHRM1 | 4280619 | 11231 | -0.004 | 0.1390 | No | ||
| 97 | OTX2 | 1190471 | 11235 | -0.004 | 0.1389 | No | ||
| 98 | CPNE6 | 6520338 | 11283 | -0.004 | 0.1364 | No | ||
| 99 | POU4F1 | 2260315 | 11338 | -0.005 | 0.1336 | No | ||
| 100 | RFX4 | 3710100 5340215 | 11426 | -0.005 | 0.1290 | No | ||
| 101 | FGF14 | 2630390 3520075 6770048 | 11478 | -0.005 | 0.1264 | No | ||
| 102 | ATP6V1E2 | 6840706 | 11561 | -0.005 | 0.1220 | No | ||
| 103 | NNAT | 4920053 5130253 | 11599 | -0.006 | 0.1201 | No | ||
| 104 | C1QTNF5 | 1690402 | 11609 | -0.006 | 0.1198 | No | ||
| 105 | CLDN16 | 1850286 | 11623 | -0.006 | 0.1192 | No | ||
| 106 | S100A16 | 2510546 | 11700 | -0.006 | 0.1152 | No | ||
| 107 | TGFB2 | 4920292 | 11752 | -0.006 | 0.1126 | No | ||
| 108 | EDN1 | 1770047 | 11775 | -0.006 | 0.1115 | No | ||
| 109 | FZD1 | 3140215 | 11807 | -0.006 | 0.1100 | No | ||
| 110 | FGF11 | 840292 | 11845 | -0.006 | 0.1081 | No | ||
| 111 | S100A14 | 450400 | 11855 | -0.007 | 0.1078 | No | ||
| 112 | TIMP4 | 6110433 | 12067 | -0.007 | 0.0965 | No | ||
| 113 | SYT7 | 2450528 3390500 3610687 | 12128 | -0.008 | 0.0934 | No | ||
| 114 | MFAP4 | 520181 | 12192 | -0.008 | 0.0902 | No | ||
| 115 | PFKFB1 | 2370128 | 12202 | -0.008 | 0.0898 | No | ||
| 116 | LHX6 | 2360731 3360397 | 12331 | -0.009 | 0.0831 | No | ||
| 117 | SMTN | 670563 1190438 | 12493 | -0.009 | 0.0746 | No | ||
| 118 | EIF3S1 | 6130368 6770044 | 12499 | -0.009 | 0.0745 | No | ||
| 119 | MEIS1 | 1400575 | 12634 | -0.010 | 0.0675 | No | ||
| 120 | AGER | 5860538 5910180 | 12762 | -0.011 | 0.0608 | No | ||
| 121 | RFX5 | 5420128 | 12959 | -0.012 | 0.0505 | No | ||
| 122 | BMPR2 | 5340066 6960328 | 13057 | -0.013 | 0.0455 | No | ||
| 123 | RUSC1 | 6450093 6980575 | 13381 | -0.015 | 0.0283 | No | ||
| 124 | RARB | 430139 1410138 | 13554 | -0.017 | 0.0193 | No | ||
| 125 | WNT10B | 510050 | 13648 | -0.018 | 0.0147 | No | ||
| 126 | COL11A2 | 5130341 | 13798 | -0.020 | 0.0071 | No | ||
| 127 | JARID2 | 6290538 | 13800 | -0.020 | 0.0074 | No | ||
| 128 | BTBD3 | 1980041 2510577 3190053 4200746 | 13823 | -0.020 | 0.0067 | No | ||
| 129 | TEAD3 | 2320142 | 13837 | -0.020 | 0.0064 | No | ||
| 130 | PAX2 | 6040270 7000133 | 13849 | -0.021 | 0.0063 | No | ||
| 131 | STX16 | 70315 | 14207 | -0.026 | -0.0125 | No | ||
| 132 | GRIN2D | 6620372 | 14233 | -0.027 | -0.0133 | No | ||
| 133 | CNTFR | 4480092 6200064 | 14421 | -0.030 | -0.0228 | No | ||
| 134 | TGIF2 | 3840364 4050520 | 14490 | -0.032 | -0.0258 | No | ||
| 135 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 14737 | -0.040 | -0.0382 | No | ||
| 136 | JMJD1C | 940575 2120025 | 14762 | -0.040 | -0.0386 | No | ||
| 137 | ACTN2 | 4200435 | 14786 | -0.042 | -0.0390 | No | ||
| 138 | POU3F4 | 870274 | 14877 | -0.045 | -0.0429 | No | ||
| 139 | FOXB1 | 4920270 5290463 | 14946 | -0.049 | -0.0455 | No | ||
| 140 | NTRK3 | 4050687 4070167 4610026 6100484 | 14992 | -0.052 | -0.0468 | No | ||
| 141 | PVRL1 | 2570167 5220519 5890500 3190309 | 15059 | -0.055 | -0.0492 | No | ||
| 142 | CRAT | 540020 2060364 3520148 | 15073 | -0.056 | -0.0487 | No | ||
| 143 | ITGAE | 4210632 4670239 5340253 5420746 5690154 5700685 6770154 | 15630 | -0.103 | -0.0766 | No | ||
| 144 | TOP1 | 770471 4060632 6650324 | 16236 | -0.176 | -0.1056 | No | ||
| 145 | ZMYND17 | 840500 | 16431 | -0.210 | -0.1115 | No | ||
| 146 | ZHX2 | 2900452 | 16790 | -0.289 | -0.1247 | No | ||
| 147 | PTPN7 | 3450110 | 16799 | -0.293 | -0.1188 | No | ||
| 148 | ALDH4A1 | 2450450 | 16813 | -0.297 | -0.1131 | No | ||
| 149 | CHD2 | 4070039 | 16918 | -0.325 | -0.1117 | No | ||
| 150 | ST5 | 3780204 | 16931 | -0.328 | -0.1052 | No | ||
| 151 | SIPA1 | 5220687 | 16935 | -0.330 | -0.0983 | No | ||
| 152 | PTPN6 | 4230128 | 17006 | -0.350 | -0.0945 | No | ||
| 153 | CSDA | 2470092 3060053 4780056 | 17011 | -0.351 | -0.0871 | No | ||
| 154 | SLC4A2 | 2690114 | 17146 | -0.399 | -0.0858 | No | ||
| 155 | BCL9L | 6940053 | 17235 | -0.427 | -0.0813 | No | ||
| 156 | CDK5 | 940348 | 17311 | -0.458 | -0.0754 | No | ||
| 157 | BAZ2A | 730184 | 17621 | -0.608 | -0.0790 | No | ||
| 158 | RAI1 | 110377 5340670 | 17641 | -0.617 | -0.0667 | No | ||
| 159 | DOCK9 | 5080037 | 17729 | -0.672 | -0.0569 | No | ||
| 160 | ASB2 | 4760168 | 17748 | -0.683 | -0.0431 | No | ||
| 161 | PDLIM2 | 2450603 2810551 3830040 4670164 | 17777 | -0.700 | -0.0295 | No | ||
| 162 | KLF7 | 50390 | 18012 | -0.886 | -0.0230 | No | ||
| 163 | AGPAT4 | 6040079 | 18093 | -0.980 | -0.0062 | No | ||
| 164 | ANKMY2 | 780600 2230440 | 18386 | -1.593 | 0.0125 | No |