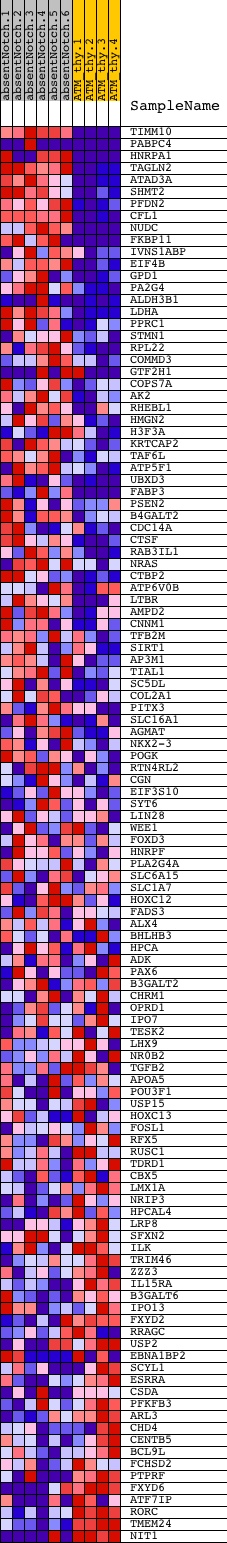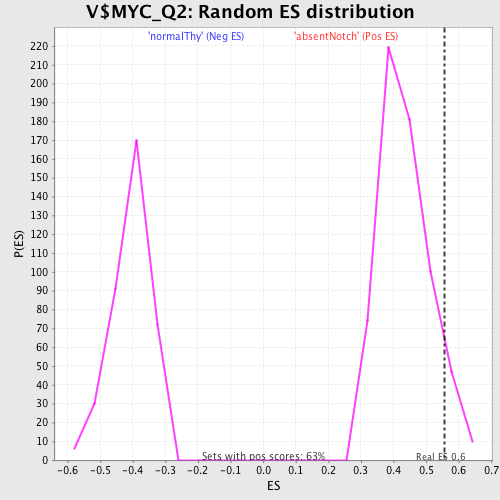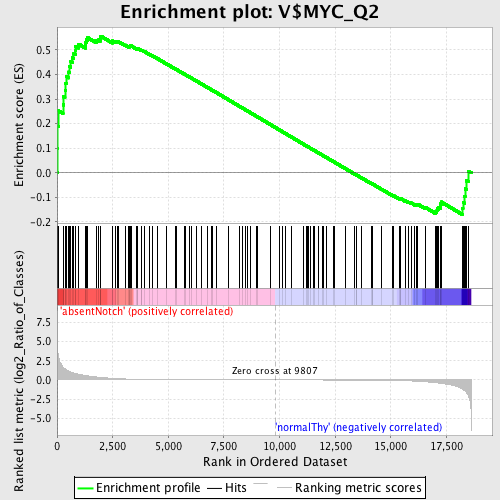
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | 0.55607766 |
| Normalized Enrichment Score (NES) | 1.2820873 |
| Nominal p-value | 0.07131537 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TIMM10 | 610070 1450019 | 12 | 4.552 | 0.0982 | Yes | ||
| 2 | PABPC4 | 1990170 6760270 5390138 | 20 | 4.232 | 0.1897 | Yes | ||
| 3 | HNRPA1 | 4670398 5050687 | 71 | 2.999 | 0.2521 | Yes | ||
| 4 | TAGLN2 | 4560121 | 267 | 1.625 | 0.2769 | Yes | ||
| 5 | ATAD3A | 4050288 | 292 | 1.547 | 0.3092 | Yes | ||
| 6 | SHMT2 | 1850180 6040010 | 355 | 1.427 | 0.3368 | Yes | ||
| 7 | PFDN2 | 4230156 | 382 | 1.369 | 0.3651 | Yes | ||
| 8 | CFL1 | 2340735 | 431 | 1.297 | 0.3907 | Yes | ||
| 9 | NUDC | 4780025 | 524 | 1.127 | 0.4102 | Yes | ||
| 10 | FKBP11 | 3170575 | 552 | 1.094 | 0.4325 | Yes | ||
| 11 | IVNS1ABP | 4760601 6520113 | 615 | 1.022 | 0.4513 | Yes | ||
| 12 | EIF4B | 5390563 5270577 | 688 | 0.933 | 0.4677 | Yes | ||
| 13 | GPD1 | 2480095 | 747 | 0.885 | 0.4838 | Yes | ||
| 14 | PA2G4 | 2030463 | 823 | 0.827 | 0.4977 | Yes | ||
| 15 | ALDH3B1 | 4210010 6940403 | 834 | 0.821 | 0.5150 | Yes | ||
| 16 | LDHA | 2190594 | 973 | 0.735 | 0.5235 | Yes | ||
| 17 | PPRC1 | 1740441 | 1271 | 0.572 | 0.5198 | Yes | ||
| 18 | STMN1 | 1990717 | 1289 | 0.561 | 0.5311 | Yes | ||
| 19 | RPL22 | 4070070 | 1319 | 0.549 | 0.5414 | Yes | ||
| 20 | COMMD3 | 2510259 | 1377 | 0.523 | 0.5497 | Yes | ||
| 21 | GTF2H1 | 2570746 2650148 | 1763 | 0.390 | 0.5374 | Yes | ||
| 22 | COPS7A | 3940458 7000022 | 1848 | 0.360 | 0.5406 | Yes | ||
| 23 | AK2 | 4590458 4760195 6110097 | 1935 | 0.334 | 0.5432 | Yes | ||
| 24 | RHEBL1 | 2810576 | 1959 | 0.327 | 0.5491 | Yes | ||
| 25 | HMGN2 | 3140091 | 1962 | 0.326 | 0.5561 | Yes | ||
| 26 | H3F3A | 2900086 | 2485 | 0.208 | 0.5324 | No | ||
| 27 | KRTCAP2 | 5220040 | 2490 | 0.206 | 0.5366 | No | ||
| 28 | TAF6L | 1850056 | 2630 | 0.179 | 0.5330 | No | ||
| 29 | ATP5F1 | 7000725 | 2708 | 0.167 | 0.5325 | No | ||
| 30 | UBXD3 | 5050193 | 2750 | 0.161 | 0.5338 | No | ||
| 31 | FABP3 | 6860452 | 3055 | 0.123 | 0.5200 | No | ||
| 32 | PSEN2 | 130382 | 3230 | 0.104 | 0.5128 | No | ||
| 33 | B4GALT2 | 780037 | 3257 | 0.101 | 0.5136 | No | ||
| 34 | CDC14A | 4050132 | 3274 | 0.100 | 0.5149 | No | ||
| 35 | CTSF | 6100021 | 3285 | 0.099 | 0.5165 | No | ||
| 36 | RAB3IL1 | 380685 5420176 | 3325 | 0.096 | 0.5165 | No | ||
| 37 | NRAS | 6900577 | 3584 | 0.077 | 0.5042 | No | ||
| 38 | CTBP2 | 430309 3710079 | 3596 | 0.076 | 0.5053 | No | ||
| 39 | ATP6V0B | 2810112 | 3618 | 0.075 | 0.5058 | No | ||
| 40 | LTBR | 2450341 | 3790 | 0.064 | 0.4979 | No | ||
| 41 | AMPD2 | 630093 1170070 | 3791 | 0.064 | 0.4993 | No | ||
| 42 | CNNM1 | 2190433 | 3911 | 0.058 | 0.4942 | No | ||
| 43 | TFB2M | 3830161 4670059 4760184 | 4138 | 0.048 | 0.4830 | No | ||
| 44 | SIRT1 | 1190731 | 4282 | 0.043 | 0.4762 | No | ||
| 45 | AP3M1 | 5340433 | 4527 | 0.036 | 0.4638 | No | ||
| 46 | TIAL1 | 4150048 6510605 | 4920 | 0.027 | 0.4432 | No | ||
| 47 | SC5DL | 6650390 | 5326 | 0.021 | 0.4217 | No | ||
| 48 | COL2A1 | 460446 3940500 | 5364 | 0.021 | 0.4202 | No | ||
| 49 | PITX3 | 3130162 | 5703 | 0.017 | 0.4023 | No | ||
| 50 | SLC16A1 | 50433 | 5769 | 0.017 | 0.3991 | No | ||
| 51 | AGMAT | 6200154 | 5953 | 0.015 | 0.3896 | No | ||
| 52 | NKX2-3 | 1990736 6940088 | 6032 | 0.015 | 0.3857 | No | ||
| 53 | POGK | 5290315 | 6284 | 0.013 | 0.3724 | No | ||
| 54 | RTN4RL2 | 6450609 | 6497 | 0.012 | 0.3612 | No | ||
| 55 | CGN | 5890139 6370167 6400537 | 6781 | 0.010 | 0.3461 | No | ||
| 56 | EIF3S10 | 1190079 3840176 | 6955 | 0.009 | 0.3369 | No | ||
| 57 | SYT6 | 2510280 3850128 4540064 | 6997 | 0.009 | 0.3349 | No | ||
| 58 | LIN28 | 6590672 | 7157 | 0.008 | 0.3265 | No | ||
| 59 | WEE1 | 3390070 | 7696 | 0.006 | 0.2975 | No | ||
| 60 | FOXD3 | 6550156 | 8199 | 0.005 | 0.2705 | No | ||
| 61 | HNRPF | 670138 | 8354 | 0.004 | 0.2623 | No | ||
| 62 | PLA2G4A | 6380364 | 8462 | 0.004 | 0.2566 | No | ||
| 63 | SLC6A15 | 5290176 6040017 | 8575 | 0.004 | 0.2506 | No | ||
| 64 | SLC1A7 | 1990181 | 8710 | 0.003 | 0.2434 | No | ||
| 65 | HOXC12 | 5270411 | 8967 | 0.002 | 0.2296 | No | ||
| 66 | FADS3 | 3120079 | 9008 | 0.002 | 0.2275 | No | ||
| 67 | ALX4 | 3870019 | 9569 | 0.001 | 0.1973 | No | ||
| 68 | BHLHB3 | 3450438 | 10001 | -0.001 | 0.1740 | No | ||
| 69 | HPCA | 6860010 | 10122 | -0.001 | 0.1675 | No | ||
| 70 | ADK | 380338 520180 5270524 | 10256 | -0.001 | 0.1603 | No | ||
| 71 | PAX6 | 1190025 | 10550 | -0.002 | 0.1445 | No | ||
| 72 | B3GALT2 | 430136 | 11078 | -0.004 | 0.1161 | No | ||
| 73 | CHRM1 | 4280619 | 11231 | -0.004 | 0.1080 | No | ||
| 74 | OPRD1 | 4280138 | 11248 | -0.004 | 0.1072 | No | ||
| 75 | IPO7 | 2190746 | 11285 | -0.004 | 0.1054 | No | ||
| 76 | TESK2 | 6520524 | 11378 | -0.005 | 0.1005 | No | ||
| 77 | LHX9 | 2350112 | 11515 | -0.005 | 0.0933 | No | ||
| 78 | NR0B2 | 2470692 | 11569 | -0.005 | 0.0905 | No | ||
| 79 | TGFB2 | 4920292 | 11752 | -0.006 | 0.0808 | No | ||
| 80 | APOA5 | 6130471 | 11913 | -0.007 | 0.0723 | No | ||
| 81 | POU3F1 | 3710022 | 11951 | -0.007 | 0.0705 | No | ||
| 82 | USP15 | 610592 3520504 | 12087 | -0.008 | 0.0634 | No | ||
| 83 | HOXC13 | 4070722 | 12441 | -0.009 | 0.0445 | No | ||
| 84 | FOSL1 | 430021 | 12472 | -0.009 | 0.0430 | No | ||
| 85 | RFX5 | 5420128 | 12959 | -0.012 | 0.0170 | No | ||
| 86 | RUSC1 | 6450093 6980575 | 13381 | -0.015 | -0.0054 | No | ||
| 87 | TDRD1 | 4230301 6110750 | 13453 | -0.016 | -0.0089 | No | ||
| 88 | CBX5 | 3830072 6290167 | 13476 | -0.016 | -0.0097 | No | ||
| 89 | LMX1A | 2810239 5690324 | 13678 | -0.018 | -0.0202 | No | ||
| 90 | NRIP3 | 1340372 | 14131 | -0.025 | -0.0441 | No | ||
| 91 | HPCAL4 | 2600484 | 14159 | -0.025 | -0.0450 | No | ||
| 92 | LRP8 | 3610746 5360035 | 14178 | -0.026 | -0.0454 | No | ||
| 93 | SFXN2 | 2650563 3140095 3830762 3870398 | 14573 | -0.034 | -0.0660 | No | ||
| 94 | ILK | 870601 4480180 | 15077 | -0.057 | -0.0919 | No | ||
| 95 | TRIM46 | 3520609 4590010 | 15140 | -0.061 | -0.0940 | No | ||
| 96 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 15397 | -0.080 | -0.1061 | No | ||
| 97 | IL15RA | 2570270 3190711 6100091 6100270 | 15418 | -0.082 | -0.1054 | No | ||
| 98 | B3GALT6 | 50195 | 15444 | -0.084 | -0.1049 | No | ||
| 99 | IPO13 | 3870088 | 15655 | -0.106 | -0.1139 | No | ||
| 100 | FXYD2 | 1400280 2350719 3520184 7040377 | 15799 | -0.121 | -0.1191 | No | ||
| 101 | RRAGC | 5910348 | 15918 | -0.132 | -0.1226 | No | ||
| 102 | USP2 | 1190292 1240253 4850035 | 16083 | -0.156 | -0.1280 | No | ||
| 103 | EBNA1BP2 | 50369 1400433 6040019 | 16145 | -0.164 | -0.1278 | No | ||
| 104 | SCYL1 | 5050717 | 16201 | -0.172 | -0.1270 | No | ||
| 105 | ESRRA | 6550242 | 16573 | -0.242 | -0.1418 | No | ||
| 106 | CSDA | 2470092 3060053 4780056 | 17011 | -0.351 | -0.1578 | No | ||
| 107 | PFKFB3 | 630706 | 17072 | -0.370 | -0.1530 | No | ||
| 108 | ARL3 | 5690725 | 17087 | -0.378 | -0.1456 | No | ||
| 109 | CHD4 | 5420059 6130338 6380717 | 17136 | -0.395 | -0.1396 | No | ||
| 110 | CENTB5 | 5690128 | 17229 | -0.425 | -0.1353 | No | ||
| 111 | BCL9L | 6940053 | 17235 | -0.427 | -0.1263 | No | ||
| 112 | FCHSD2 | 5720092 | 17265 | -0.438 | -0.1184 | No | ||
| 113 | PTPRF | 1770528 3190044 | 18211 | -1.146 | -0.1446 | No | ||
| 114 | FXYD6 | 3870164 | 18246 | -1.202 | -0.1203 | No | ||
| 115 | ATF7IP | 2690176 6770021 | 18306 | -1.342 | -0.0944 | No | ||
| 116 | RORC | 1740121 | 18341 | -1.439 | -0.0650 | No | ||
| 117 | TMEM24 | 5860039 | 18395 | -1.644 | -0.0321 | No | ||
| 118 | NIT1 | 1230338 3940671 | 18488 | -2.027 | 0.0069 | No |