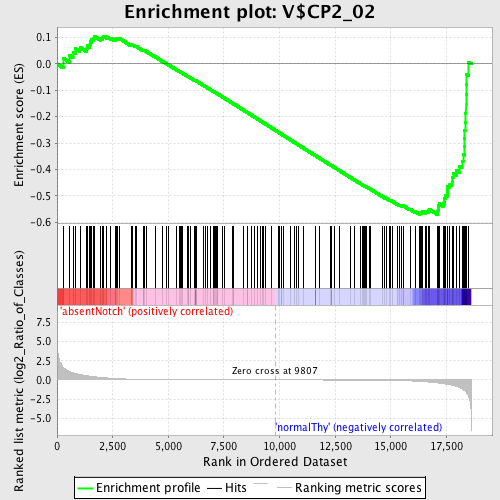
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$CP2_02 |
| Enrichment Score (ES) | -0.57118094 |
| Normalized Enrichment Score (NES) | -1.4516453 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.24437967 |
| FWER p-Value | 0.945 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDIT4 | 2570408 | 281 | 1.582 | 0.0215 | No | ||
| 2 | NR1D1 | 2360471 770746 6590204 | 543 | 1.102 | 0.0329 | No | ||
| 3 | PRKACA | 2640731 4050048 | 715 | 0.911 | 0.0448 | No | ||
| 4 | ALDH3B1 | 4210010 6940403 | 834 | 0.821 | 0.0574 | No | ||
| 5 | PHPT1 | 7050537 | 1028 | 0.697 | 0.0632 | No | ||
| 6 | KLHDC3 | 3830717 | 1341 | 0.538 | 0.0587 | No | ||
| 7 | RASSF1 | 2760364 4850725 | 1376 | 0.523 | 0.0690 | No | ||
| 8 | GRB7 | 2100471 | 1477 | 0.484 | 0.0748 | No | ||
| 9 | KCTD15 | 2810735 | 1516 | 0.469 | 0.0837 | No | ||
| 10 | PPP1R16B | 3520300 | 1559 | 0.450 | 0.0918 | No | ||
| 11 | NISCH | 4850301 | 1622 | 0.429 | 0.0984 | No | ||
| 12 | MRPL14 | 2370168 | 1685 | 0.409 | 0.1045 | No | ||
| 13 | H3F3B | 1410300 | 1955 | 0.329 | 0.0976 | No | ||
| 14 | MORF4L2 | 6450133 | 2024 | 0.312 | 0.1011 | No | ||
| 15 | CS | 5080600 | 2070 | 0.301 | 0.1057 | No | ||
| 16 | RYR1 | 2510114 3130408 3710086 | 2238 | 0.255 | 0.1026 | No | ||
| 17 | HTATIP | 5360739 | 2421 | 0.219 | 0.0978 | No | ||
| 18 | TGFBR2 | 1780711 1980537 6550398 | 2610 | 0.183 | 0.0918 | No | ||
| 19 | LHX2 | 3610463 | 2645 | 0.178 | 0.0941 | No | ||
| 20 | CAMK2G | 630097 | 2670 | 0.174 | 0.0969 | No | ||
| 21 | GMPPB | 4610538 | 2724 | 0.165 | 0.0978 | No | ||
| 22 | FKBP2 | 2320025 | 2799 | 0.153 | 0.0974 | No | ||
| 23 | PRKAG1 | 6450091 | 3321 | 0.096 | 0.0714 | No | ||
| 24 | NR2F1 | 4120528 360402 | 3333 | 0.095 | 0.0730 | No | ||
| 25 | RHBDF1 | 380538 | 3391 | 0.091 | 0.0720 | No | ||
| 26 | SHB | 4920494 | 3522 | 0.080 | 0.0668 | No | ||
| 27 | RBBP6 | 2320129 | 3567 | 0.078 | 0.0662 | No | ||
| 28 | CNTN6 | 2630711 | 3872 | 0.060 | 0.0512 | No | ||
| 29 | EPN3 | 2350592 | 3876 | 0.060 | 0.0524 | No | ||
| 30 | PPP2R5C | 3310121 6650672 | 3930 | 0.057 | 0.0508 | No | ||
| 31 | TUFT1 | 540047 2370010 | 4035 | 0.052 | 0.0464 | No | ||
| 32 | DSCAM | 1780050 2450731 2810438 | 4405 | 0.039 | 0.0273 | No | ||
| 33 | MMP11 | 1980619 | 4747 | 0.030 | 0.0096 | No | ||
| 34 | GPC3 | 6180735 | 4927 | 0.027 | 0.0005 | No | ||
| 35 | ETV1 | 70735 2940603 5080463 | 5007 | 0.026 | -0.0032 | No | ||
| 36 | RTN1 | 4150452 4850176 | 5345 | 0.021 | -0.0210 | No | ||
| 37 | FGF9 | 1050195 | 5517 | 0.019 | -0.0298 | No | ||
| 38 | STAC2 | 7100022 | 5535 | 0.019 | -0.0303 | No | ||
| 39 | SLC25A28 | 6590600 | 5579 | 0.018 | -0.0322 | No | ||
| 40 | FOXP2 | 3520561 4150372 4760524 | 5637 | 0.018 | -0.0349 | No | ||
| 41 | STARD13 | 540722 | 5844 | 0.016 | -0.0457 | No | ||
| 42 | COX8C | 4670154 | 5888 | 0.016 | -0.0476 | No | ||
| 43 | DHH | 2120433 | 5923 | 0.015 | -0.0491 | No | ||
| 44 | TBX2 | 1990563 | 5988 | 0.015 | -0.0523 | No | ||
| 45 | TNNC1 | 1990575 | 6161 | 0.014 | -0.0613 | No | ||
| 46 | FEV | 840369 5860139 | 6214 | 0.013 | -0.0638 | No | ||
| 47 | GPRC5C | 1580372 | 6216 | 0.013 | -0.0635 | No | ||
| 48 | NRIP2 | 5390288 | 6217 | 0.013 | -0.0632 | No | ||
| 49 | MAPRE3 | 7050504 | 6232 | 0.013 | -0.0637 | No | ||
| 50 | NFATC4 | 2470735 | 6252 | 0.013 | -0.0644 | No | ||
| 51 | RNF121 | 110079 1580279 | 6270 | 0.013 | -0.0650 | No | ||
| 52 | TLX3 | 510619 | 6597 | 0.011 | -0.0824 | No | ||
| 53 | EFNA3 | 6370390 | 6659 | 0.011 | -0.0855 | No | ||
| 54 | DCX | 130075 5360025 7050673 | 6737 | 0.010 | -0.0894 | No | ||
| 55 | PFTK1 | 1780181 | 6876 | 0.009 | -0.0967 | No | ||
| 56 | SRMS | 5360402 | 7013 | 0.009 | -0.1038 | No | ||
| 57 | KCNE3 | 5550195 | 7024 | 0.009 | -0.1042 | No | ||
| 58 | HPCAL1 | 1980129 2350348 | 7052 | 0.009 | -0.1054 | No | ||
| 59 | GSH1 | 2760242 | 7103 | 0.009 | -0.1079 | No | ||
| 60 | CITED1 | 2350670 | 7176 | 0.008 | -0.1117 | No | ||
| 61 | KIF1C | 2480484 | 7189 | 0.008 | -0.1121 | No | ||
| 62 | GNB3 | 1230167 | 7194 | 0.008 | -0.1121 | No | ||
| 63 | LYN | 6040600 | 7424 | 0.007 | -0.1244 | No | ||
| 64 | HSPA1L | 4010538 | 7540 | 0.007 | -0.1305 | No | ||
| 65 | PODN | 3710068 | 7878 | 0.006 | -0.1486 | No | ||
| 66 | MID2 | 4120309 | 7926 | 0.005 | -0.1510 | No | ||
| 67 | FOXL2 | 5670537 | 7946 | 0.005 | -0.1519 | No | ||
| 68 | CALD1 | 1770129 1940397 | 8364 | 0.004 | -0.1744 | No | ||
| 69 | CREB1 | 1500717 2230358 3610600 6550601 | 8539 | 0.004 | -0.1837 | No | ||
| 70 | SLC26A9 | 3440722 5340088 7510093 | 8546 | 0.004 | -0.1840 | No | ||
| 71 | UPK2 | 60176 | 8719 | 0.003 | -0.1932 | No | ||
| 72 | BRUNOL6 | 6840373 | 8865 | 0.003 | -0.2010 | No | ||
| 73 | SLC2A9 | 3290301 | 9026 | 0.002 | -0.2096 | No | ||
| 74 | AQP6 | 510736 1570484 | 9134 | 0.002 | -0.2154 | No | ||
| 75 | FRMD3 | 3440731 | 9227 | 0.002 | -0.2203 | No | ||
| 76 | TBC1D5 | 5360619 6130603 | 9297 | 0.001 | -0.2241 | No | ||
| 77 | XPNPEP1 | 2640750 3850315 6200242 | 9367 | 0.001 | -0.2278 | No | ||
| 78 | ODC1 | 5670168 | 9621 | 0.001 | -0.2415 | No | ||
| 79 | PCDHB4 | 670184 | 9933 | -0.000 | -0.2583 | No | ||
| 80 | ARHGAP5 | 2510619 3360035 | 9989 | -0.001 | -0.2613 | No | ||
| 81 | ADRA1A | 1230446 2260390 | 10080 | -0.001 | -0.2661 | No | ||
| 82 | HOXB6 | 4210739 | 10097 | -0.001 | -0.2670 | No | ||
| 83 | NRXN3 | 1400546 3360471 | 10155 | -0.001 | -0.2700 | No | ||
| 84 | ARX | 6900504 | 10474 | -0.002 | -0.2872 | No | ||
| 85 | PTHLH | 5290739 | 10686 | -0.002 | -0.2986 | No | ||
| 86 | RAB10 | 2360008 5890020 | 10751 | -0.003 | -0.3020 | No | ||
| 87 | PAK7 | 3830164 | 10833 | -0.003 | -0.3063 | No | ||
| 88 | BMP4 | 380113 | 11085 | -0.004 | -0.3198 | No | ||
| 89 | DSC2 | 840484 | 11598 | -0.006 | -0.3474 | No | ||
| 90 | NNAT | 4920053 5130253 | 11599 | -0.006 | -0.3473 | No | ||
| 91 | C1QTNF5 | 1690402 | 11609 | -0.006 | -0.3477 | No | ||
| 92 | FOXA1 | 3710609 | 11619 | -0.006 | -0.3480 | No | ||
| 93 | BDNF | 2940128 3520368 | 11795 | -0.006 | -0.3574 | No | ||
| 94 | HIF1A | 5670605 | 12286 | -0.008 | -0.3837 | No | ||
| 95 | PITX2 | 870537 2690139 | 12317 | -0.009 | -0.3851 | No | ||
| 96 | P518 | 3610435 | 12326 | -0.009 | -0.3854 | No | ||
| 97 | PAPPA | 4230463 | 12352 | -0.009 | -0.3865 | No | ||
| 98 | CAPN6 | 1740168 | 12459 | -0.009 | -0.3921 | No | ||
| 99 | TRERF1 | 6370017 6940138 | 12707 | -0.011 | -0.4052 | No | ||
| 100 | PLXNA3 | 7050164 | 13167 | -0.014 | -0.4297 | No | ||
| 101 | MOGAT2 | 60139 | 13354 | -0.015 | -0.4395 | No | ||
| 102 | WNT10B | 510050 | 13648 | -0.018 | -0.4549 | No | ||
| 103 | TXNL5 | 1190609 1400164 | 13710 | -0.019 | -0.4578 | No | ||
| 104 | GNAQ | 430670 4210131 5900736 | 13759 | -0.019 | -0.4599 | No | ||
| 105 | RPL41 | 6940112 | 13805 | -0.020 | -0.4619 | No | ||
| 106 | PAX2 | 6040270 7000133 | 13849 | -0.021 | -0.4638 | No | ||
| 107 | IRX3 | 4200112 | 13886 | -0.021 | -0.4652 | No | ||
| 108 | NOL4 | 5050446 5360519 | 14046 | -0.024 | -0.4733 | No | ||
| 109 | KIF5A | 510435 | 14097 | -0.024 | -0.4754 | No | ||
| 110 | DDR1 | 2060044 5220180 | 14631 | -0.036 | -0.5035 | No | ||
| 111 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 14737 | -0.040 | -0.5083 | No | ||
| 112 | LDB2 | 5670441 6110170 | 14784 | -0.041 | -0.5098 | No | ||
| 113 | MDK | 3840020 | 14925 | -0.048 | -0.5163 | No | ||
| 114 | NTRK3 | 4050687 4070167 4610026 6100484 | 14992 | -0.052 | -0.5186 | No | ||
| 115 | BTBD14B | 3520601 4280162 | 15087 | -0.057 | -0.5224 | No | ||
| 116 | TRAF4 | 3060041 4920528 6980286 | 15313 | -0.073 | -0.5329 | No | ||
| 117 | ZNF385 | 450176 4810358 | 15405 | -0.080 | -0.5360 | No | ||
| 118 | CPNE1 | 3440131 | 15464 | -0.086 | -0.5371 | No | ||
| 119 | MNT | 4920575 | 15554 | -0.096 | -0.5397 | No | ||
| 120 | KIFAP3 | 3800524 4590082 6940075 | 15563 | -0.096 | -0.5379 | No | ||
| 121 | NEK8 | 5700180 | 15862 | -0.128 | -0.5511 | No | ||
| 122 | SCAMP5 | 5550368 6290021 | 16111 | -0.159 | -0.5608 | No | ||
| 123 | YWHAG | 3780341 | 16288 | -0.184 | -0.5661 | Yes | ||
| 124 | UBE1 | 4150139 7050463 | 16334 | -0.191 | -0.5641 | Yes | ||
| 125 | DDAH2 | 4480551 | 16392 | -0.203 | -0.5625 | Yes | ||
| 126 | CRK | 1230162 4780128 | 16406 | -0.206 | -0.5584 | Yes | ||
| 127 | TLN1 | 6590411 | 16541 | -0.235 | -0.5602 | Yes | ||
| 128 | SLC9A1 | 2650093 | 16624 | -0.253 | -0.5588 | Yes | ||
| 129 | BACH1 | 290195 | 16687 | -0.268 | -0.5559 | Yes | ||
| 130 | FLI1 | 3990142 | 16745 | -0.280 | -0.5525 | Yes | ||
| 131 | RHBDL1 | 1500736 | 17090 | -0.379 | -0.5624 | Yes | ||
| 132 | ATP10D | 4850139 | 17119 | -0.391 | -0.5548 | Yes | ||
| 133 | GSK3B | 5360348 | 17132 | -0.395 | -0.5463 | Yes | ||
| 134 | CHD4 | 5420059 6130338 6380717 | 17136 | -0.395 | -0.5373 | Yes | ||
| 135 | SLC25A14 | 4760180 | 17166 | -0.403 | -0.5295 | Yes | ||
| 136 | MAPK3 | 580161 4780035 | 17348 | -0.472 | -0.5284 | Yes | ||
| 137 | TRIM8 | 6130441 | 17410 | -0.501 | -0.5201 | Yes | ||
| 138 | MAST2 | 6200348 | 17415 | -0.502 | -0.5087 | Yes | ||
| 139 | CLDN4 | 4920739 | 17443 | -0.513 | -0.4982 | Yes | ||
| 140 | HCFC1 | 2260008 | 17532 | -0.552 | -0.4902 | Yes | ||
| 141 | LZTS2 | 2260440 | 17544 | -0.558 | -0.4778 | Yes | ||
| 142 | GLTP | 3440603 | 17562 | -0.569 | -0.4655 | Yes | ||
| 143 | NRF1 | 2650195 | 17655 | -0.626 | -0.4560 | Yes | ||
| 144 | TRPV2 | 7100348 | 17750 | -0.684 | -0.4452 | Yes | ||
| 145 | PRKCH | 5720079 | 17758 | -0.689 | -0.4296 | Yes | ||
| 146 | GABARAP | 1450286 | 17808 | -0.719 | -0.4156 | Yes | ||
| 147 | JUNB | 4230048 | 17941 | -0.818 | -0.4038 | Yes | ||
| 148 | FLOT2 | 50358 4210068 5080397 | 18102 | -0.989 | -0.3895 | Yes | ||
| 149 | NGFRAP1 | 6770195 | 18200 | -1.130 | -0.3686 | Yes | ||
| 150 | DNAJB1 | 1090041 | 18250 | -1.209 | -0.3432 | Yes | ||
| 151 | CDC42EP3 | 2480138 | 18299 | -1.335 | -0.3148 | Yes | ||
| 152 | CAP1 | 2650278 | 18305 | -1.341 | -0.2840 | Yes | ||
| 153 | FBS1 | 2570520 | 18317 | -1.366 | -0.2529 | Yes | ||
| 154 | BCL11B | 2680673 | 18339 | -1.434 | -0.2207 | Yes | ||
| 155 | DEF6 | 840593 | 18349 | -1.475 | -0.1870 | Yes | ||
| 156 | EPC1 | 1570050 5290095 6900193 | 18379 | -1.571 | -0.1521 | Yes | ||
| 157 | PPP1R9B | 3130619 | 18381 | -1.573 | -0.1157 | Yes | ||
| 158 | KCNH2 | 1170451 | 18397 | -1.648 | -0.0783 | Yes | ||
| 159 | PTCRA | 6940142 | 18404 | -1.670 | -0.0398 | Yes | ||
| 160 | DUSP6 | 5910286 7100070 | 18507 | -2.210 | 0.0059 | Yes |