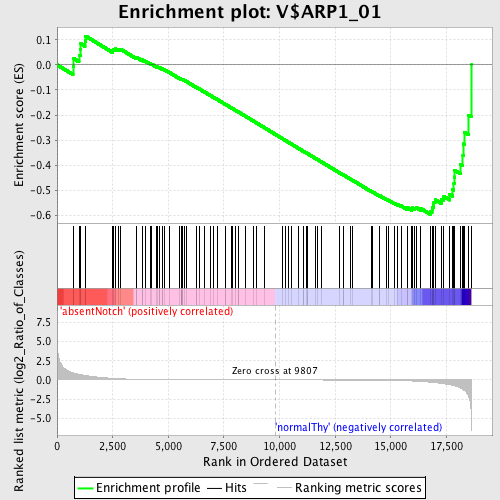
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | V$ARP1_01 |
| Enrichment Score (ES) | -0.59524524 |
| Normalized Enrichment Score (NES) | -1.4319323 |
| Nominal p-value | 0.012886598 |
| FDR q-value | 0.27705228 |
| FWER p-Value | 0.98 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ALDOA | 6290672 | 716 | 0.911 | -0.0054 | No | ||
| 2 | MRPS18B | 5570086 | 751 | 0.883 | 0.0250 | No | ||
| 3 | PMF1 | 5130273 | 984 | 0.729 | 0.0390 | No | ||
| 4 | DLX1 | 360168 | 1042 | 0.689 | 0.0611 | No | ||
| 5 | NDST2 | 6290711 | 1064 | 0.674 | 0.0846 | No | ||
| 6 | ATBF1 | 1240131 7040465 | 1254 | 0.577 | 0.0954 | No | ||
| 7 | NFIX | 2450152 | 1286 | 0.564 | 0.1144 | No | ||
| 8 | RASIP1 | 1580021 | 2482 | 0.208 | 0.0574 | No | ||
| 9 | TGFB3 | 1070041 | 2515 | 0.200 | 0.0630 | No | ||
| 10 | LTBP3 | 1190162 2650440 6520451 | 2627 | 0.180 | 0.0635 | No | ||
| 11 | PROSAPIP1 | 1340113 4670440 | 2776 | 0.158 | 0.0613 | No | ||
| 12 | IRAK1 | 4120593 | 2847 | 0.147 | 0.0629 | No | ||
| 13 | RBBP6 | 2320129 | 3567 | 0.078 | 0.0269 | No | ||
| 14 | ENSA | 6620546 | 3588 | 0.077 | 0.0286 | No | ||
| 15 | COL16A1 | 1780520 | 3834 | 0.061 | 0.0176 | No | ||
| 16 | CASK | 2340215 3290576 6770215 | 3837 | 0.061 | 0.0197 | No | ||
| 17 | GATA1 | 6400068 | 3978 | 0.055 | 0.0142 | No | ||
| 18 | ZFX | 5900400 | 4176 | 0.047 | 0.0052 | No | ||
| 19 | DLX5 | 5130519 6040021 | 4246 | 0.044 | 0.0031 | No | ||
| 20 | MAB21L2 | 1770605 | 4486 | 0.037 | -0.0084 | No | ||
| 21 | RIN1 | 510593 | 4506 | 0.036 | -0.0081 | No | ||
| 22 | ASB12 | 7040398 | 4586 | 0.034 | -0.0112 | No | ||
| 23 | RUNX3 | 3800546 | 4613 | 0.033 | -0.0113 | No | ||
| 24 | CTGF | 4540577 | 4717 | 0.031 | -0.0158 | No | ||
| 25 | HOXA3 | 3610632 6020358 | 4725 | 0.031 | -0.0150 | No | ||
| 26 | FCHSD1 | 5080050 6450025 | 4838 | 0.028 | -0.0200 | No | ||
| 27 | SLITRK6 | 7320739 | 5040 | 0.025 | -0.0300 | No | ||
| 28 | NXPH1 | 3870546 | 5490 | 0.019 | -0.0535 | No | ||
| 29 | HOXA1 | 1190524 5420142 | 5594 | 0.018 | -0.0584 | No | ||
| 30 | GPR3 | 2970452 | 5609 | 0.018 | -0.0585 | No | ||
| 31 | IL21 | 1850047 | 5610 | 0.018 | -0.0579 | No | ||
| 32 | FOXP2 | 3520561 4150372 4760524 | 5637 | 0.018 | -0.0586 | No | ||
| 33 | ESRRG | 4010181 | 5656 | 0.018 | -0.0590 | No | ||
| 34 | KIF13A | 5390750 | 5713 | 0.017 | -0.0614 | No | ||
| 35 | MYL1 | 2450707 | 5824 | 0.016 | -0.0667 | No | ||
| 36 | JPH1 | 610739 | 6272 | 0.013 | -0.0904 | No | ||
| 37 | MLLT6 | 3870168 | 6281 | 0.013 | -0.0904 | No | ||
| 38 | ARR3 | 130142 3850132 | 6392 | 0.012 | -0.0959 | No | ||
| 39 | NR5A2 | 1690180 6900050 | 6623 | 0.011 | -0.1079 | No | ||
| 40 | SLC12A5 | 1980692 | 6881 | 0.009 | -0.1214 | No | ||
| 41 | CLDN15 | 4150270 4730592 | 7015 | 0.009 | -0.1283 | No | ||
| 42 | CRYGC | 1660338 | 7205 | 0.008 | -0.1382 | No | ||
| 43 | SPARC | 1690086 | 7576 | 0.007 | -0.1580 | No | ||
| 44 | HAPLN1 | 580398 | 7584 | 0.007 | -0.1581 | No | ||
| 45 | TNRC4 | 4050156 | 7834 | 0.006 | -0.1713 | No | ||
| 46 | MMP28 | 2360242 | 7891 | 0.006 | -0.1742 | No | ||
| 47 | C1QTNF7 | 2970619 | 8031 | 0.005 | -0.1815 | No | ||
| 48 | CDX1 | 4570168 | 8135 | 0.005 | -0.1869 | No | ||
| 49 | OMD | 6110044 | 8147 | 0.005 | -0.1873 | No | ||
| 50 | HSPB8 | 540563 | 8487 | 0.004 | -0.2055 | No | ||
| 51 | HOXD4 | 5900301 | 8826 | 0.003 | -0.2236 | No | ||
| 52 | SKIL | 1090364 | 8954 | 0.002 | -0.2304 | No | ||
| 53 | ZNF205 | 6900022 | 9305 | 0.001 | -0.2492 | No | ||
| 54 | HPCA | 6860010 | 10122 | -0.001 | -0.2933 | No | ||
| 55 | HTR1B | 5340133 | 10252 | -0.001 | -0.3002 | No | ||
| 56 | KIRREL2 | 130168 3440097 | 10404 | -0.002 | -0.3083 | No | ||
| 57 | MNAT1 | 730292 2350364 | 10541 | -0.002 | -0.3156 | No | ||
| 58 | PROP1 | 1980008 | 10843 | -0.003 | -0.3317 | No | ||
| 59 | SERPINC1 | 1780717 | 11071 | -0.004 | -0.3439 | No | ||
| 60 | B3GALT2 | 430136 | 11078 | -0.004 | -0.3440 | No | ||
| 61 | DNAJC5B | 60368 | 11199 | -0.004 | -0.3504 | No | ||
| 62 | OTX2 | 1190471 | 11235 | -0.004 | -0.3521 | No | ||
| 63 | DMD | 1740041 3990332 | 11250 | -0.004 | -0.3527 | No | ||
| 64 | C1QTNF5 | 1690402 | 11609 | -0.006 | -0.3718 | No | ||
| 65 | FGF20 | 5690601 | 11715 | -0.006 | -0.3773 | No | ||
| 66 | BMP5 | 4670048 | 11864 | -0.007 | -0.3851 | No | ||
| 67 | TRERF1 | 6370017 6940138 | 12707 | -0.011 | -0.4302 | No | ||
| 68 | GPR124 | 510292 6290047 | 12882 | -0.012 | -0.4391 | No | ||
| 69 | AQP7 | 2760576 2810072 2940722 | 12891 | -0.012 | -0.4391 | No | ||
| 70 | APOM | 2340010 | 13205 | -0.014 | -0.4555 | No | ||
| 71 | CACNA1G | 3060161 4810706 5550722 | 13288 | -0.014 | -0.4594 | No | ||
| 72 | CYP2E1 | 4480209 | 14143 | -0.025 | -0.5047 | No | ||
| 73 | CTLA4 | 6590537 | 14189 | -0.026 | -0.5061 | No | ||
| 74 | TGIF2 | 3840364 4050520 | 14490 | -0.032 | -0.5212 | No | ||
| 75 | MYOCD | 2650064 3440403 6220121 | 14503 | -0.032 | -0.5207 | No | ||
| 76 | CFL2 | 380239 6200368 | 14783 | -0.041 | -0.5342 | No | ||
| 77 | POU3F4 | 870274 | 14877 | -0.045 | -0.5376 | No | ||
| 78 | NLK | 2030010 2450041 | 15169 | -0.062 | -0.5510 | No | ||
| 79 | TRAF4 | 3060041 4920528 6980286 | 15313 | -0.073 | -0.5561 | No | ||
| 80 | CPNE1 | 3440131 | 15464 | -0.086 | -0.5611 | No | ||
| 81 | PITPNC1 | 3990017 | 15734 | -0.114 | -0.5715 | No | ||
| 82 | PDGFB | 3060440 6370008 | 15750 | -0.115 | -0.5681 | No | ||
| 83 | ATP2C1 | 2630446 6520253 | 15946 | -0.136 | -0.5737 | No | ||
| 84 | ELF4 | 4850193 | 15955 | -0.137 | -0.5691 | No | ||
| 85 | SLC7A8 | 380136 5690672 | 16078 | -0.155 | -0.5700 | No | ||
| 86 | STAR | 7040670 | 16134 | -0.162 | -0.5671 | No | ||
| 87 | NR6A1 | 4010347 | 16354 | -0.196 | -0.5717 | No | ||
| 88 | ZHX2 | 2900452 | 16790 | -0.289 | -0.5847 | Yes | ||
| 89 | RALY | 940427 1740066 | 16852 | -0.307 | -0.5768 | Yes | ||
| 90 | AP1B1 | 4290373 | 16864 | -0.309 | -0.5661 | Yes | ||
| 91 | CHD2 | 4070039 | 16918 | -0.325 | -0.5571 | Yes | ||
| 92 | SIPA1 | 5220687 | 16935 | -0.330 | -0.5459 | Yes | ||
| 93 | PLK3 | 2640592 | 16988 | -0.346 | -0.5361 | Yes | ||
| 94 | FCHSD2 | 5720092 | 17265 | -0.438 | -0.5350 | Yes | ||
| 95 | FBXL20 | 4200064 | 17381 | -0.486 | -0.5235 | Yes | ||
| 96 | RRAGD | 670075 | 17646 | -0.622 | -0.5150 | Yes | ||
| 97 | PKP3 | 3710048 | 17779 | -0.702 | -0.4965 | Yes | ||
| 98 | PIK3CG | 5890110 | 17834 | -0.735 | -0.4726 | Yes | ||
| 99 | POLD4 | 2320082 | 17847 | -0.744 | -0.4461 | Yes | ||
| 100 | DBP | 4200270 | 17863 | -0.755 | -0.4194 | Yes | ||
| 101 | RNF34 | 3800332 4590717 5670138 | 18132 | -1.024 | -0.3965 | Yes | ||
| 102 | EMP1 | 4120438 | 18241 | -1.190 | -0.3589 | Yes | ||
| 103 | NFE2 | 3710373 | 18260 | -1.239 | -0.3146 | Yes | ||
| 104 | ATF7IP | 2690176 6770021 | 18306 | -1.342 | -0.2681 | Yes | ||
| 105 | CDK5R1 | 3870161 | 18498 | -2.123 | -0.2009 | Yes | ||
| 106 | HRBL | 1850102 3830253 4780176 | 18611 | -5.676 | 0.0003 | Yes |

