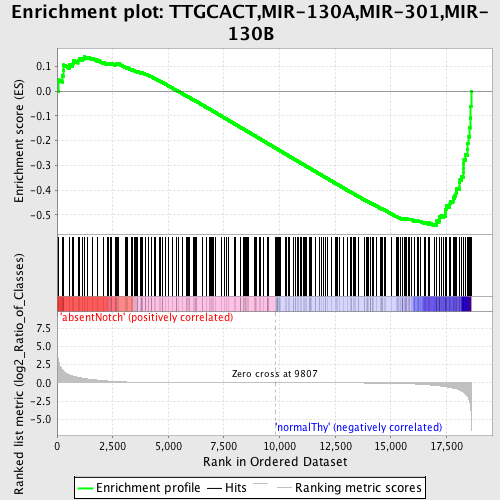
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | TTGCACT,MIR-130A,MIR-301,MIR-130B |
| Enrichment Score (ES) | -0.5449053 |
| Normalized Enrichment Score (NES) | -1.4544603 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2519002 |
| FWER p-Value | 0.94 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CHST1 | 50601 | 46 | 3.332 | 0.0466 | No | ||
| 2 | INHBB | 2680092 | 227 | 1.782 | 0.0631 | No | ||
| 3 | SLMAP | 3120093 4760128 5390528 | 271 | 1.613 | 0.0845 | No | ||
| 4 | GJA1 | 5220731 | 295 | 1.543 | 0.1060 | No | ||
| 5 | RAB34 | 770411 | 558 | 1.087 | 0.1078 | No | ||
| 6 | MBNL1 | 2640762 7100048 | 713 | 0.914 | 0.1129 | No | ||
| 7 | FASTK | 2690053 4280288 4610551 6370292 | 733 | 0.896 | 0.1250 | No | ||
| 8 | SKP1A | 2450102 | 939 | 0.756 | 0.1250 | No | ||
| 9 | SMOC2 | 1170170 | 988 | 0.724 | 0.1331 | No | ||
| 10 | CUL3 | 1850520 | 1143 | 0.637 | 0.1341 | No | ||
| 11 | YTHDF2 | 4060332 4610279 | 1219 | 0.596 | 0.1388 | No | ||
| 12 | RASSF1 | 2760364 4850725 | 1376 | 0.523 | 0.1380 | No | ||
| 13 | ABCB7 | 4150324 | 1591 | 0.440 | 0.1328 | No | ||
| 14 | CYLD | 6590079 | 1812 | 0.370 | 0.1263 | No | ||
| 15 | SFMBT1 | 2360450 2370070 | 2106 | 0.288 | 0.1146 | No | ||
| 16 | SNX2 | 2470068 3360364 6510064 | 2246 | 0.254 | 0.1107 | No | ||
| 17 | UBE2D2 | 4590671 6510446 | 2298 | 0.244 | 0.1115 | No | ||
| 18 | SPG20 | 3360202 | 2382 | 0.226 | 0.1104 | No | ||
| 19 | TTYH3 | 2100082 | 2444 | 0.215 | 0.1102 | No | ||
| 20 | TRPS1 | 870100 | 2453 | 0.213 | 0.1129 | No | ||
| 21 | TGFBR2 | 1780711 1980537 6550398 | 2610 | 0.183 | 0.1071 | No | ||
| 22 | BLCAP | 3990253 | 2640 | 0.179 | 0.1082 | No | ||
| 23 | MIB1 | 3800537 | 2654 | 0.176 | 0.1101 | No | ||
| 24 | TPP1 | 3990446 | 2684 | 0.172 | 0.1110 | No | ||
| 25 | FBXO28 | 1340358 4730100 | 2713 | 0.167 | 0.1120 | No | ||
| 26 | RAP2C | 1690132 | 2779 | 0.157 | 0.1107 | No | ||
| 27 | TMEM32 | 1170452 | 3081 | 0.120 | 0.0961 | No | ||
| 28 | HABP4 | 840487 | 3132 | 0.114 | 0.0951 | No | ||
| 29 | STC1 | 360161 | 3185 | 0.109 | 0.0938 | No | ||
| 30 | LRP4 | 1740253 | 3344 | 0.094 | 0.0866 | No | ||
| 31 | SMAD5 | 5700497 | 3366 | 0.092 | 0.0868 | No | ||
| 32 | ACSL1 | 2900520 | 3475 | 0.083 | 0.0822 | No | ||
| 33 | NME7 | 60170 1230551 2850333 4200102 4560411 5090647 | 3524 | 0.080 | 0.0808 | No | ||
| 34 | RHOT1 | 6760692 | 3586 | 0.077 | 0.0786 | No | ||
| 35 | MID1IP1 | 2570739 | 3601 | 0.076 | 0.0789 | No | ||
| 36 | CLTC | 6590458 | 3605 | 0.076 | 0.0799 | No | ||
| 37 | KIT | 7040095 | 3619 | 0.075 | 0.0803 | No | ||
| 38 | CSF1 | 3120400 | 3729 | 0.068 | 0.0753 | No | ||
| 39 | ARID4B | 730754 6660026 | 3777 | 0.065 | 0.0737 | No | ||
| 40 | SOX21 | 1050110 4670041 6660338 | 3799 | 0.064 | 0.0735 | No | ||
| 41 | PDGFRA | 2940332 | 3806 | 0.063 | 0.0741 | No | ||
| 42 | EPC2 | 2470095 | 3830 | 0.062 | 0.0738 | No | ||
| 43 | ARFIP1 | 3130601 | 3842 | 0.061 | 0.0741 | No | ||
| 44 | ACVR1 | 6840671 | 3959 | 0.056 | 0.0686 | No | ||
| 45 | SH3D19 | 6020520 | 3979 | 0.055 | 0.0684 | No | ||
| 46 | PPARG | 1990168 2680603 6130632 | 3994 | 0.054 | 0.0684 | No | ||
| 47 | CD69 | 380167 4730088 | 4094 | 0.050 | 0.0638 | No | ||
| 48 | PRKD3 | 5220520 5890519 | 4124 | 0.049 | 0.0629 | No | ||
| 49 | CANX | 4760402 | 4220 | 0.045 | 0.0584 | No | ||
| 50 | EDA | 6420441 | 4230 | 0.045 | 0.0586 | No | ||
| 51 | SNPH | 2680484 | 4242 | 0.044 | 0.0586 | No | ||
| 52 | ROBO2 | 450136 | 4394 | 0.040 | 0.0510 | No | ||
| 53 | PTPRM | 6370136 | 4422 | 0.038 | 0.0501 | No | ||
| 54 | RUNX3 | 3800546 | 4613 | 0.033 | 0.0402 | No | ||
| 55 | ESR1 | 4060372 5860193 | 4638 | 0.033 | 0.0394 | No | ||
| 56 | HOXA3 | 3610632 6020358 | 4725 | 0.031 | 0.0352 | No | ||
| 57 | ZFYVE9 | 6980288 7100017 | 4737 | 0.030 | 0.0350 | No | ||
| 58 | PRKAA1 | 510156 | 4753 | 0.030 | 0.0346 | No | ||
| 59 | SNAP25 | 360520 | 4885 | 0.028 | 0.0279 | No | ||
| 60 | RASD1 | 2450463 | 5027 | 0.025 | 0.0206 | No | ||
| 61 | ARRDC3 | 4610390 | 5195 | 0.023 | 0.0118 | No | ||
| 62 | RTN1 | 4150452 4850176 | 5345 | 0.021 | 0.0040 | No | ||
| 63 | SASH1 | 4070270 | 5346 | 0.021 | 0.0043 | No | ||
| 64 | NR3C2 | 2480324 6400093 | 5377 | 0.020 | 0.0030 | No | ||
| 65 | ACSL4 | 510020 1240041 3190050 4540500 5570463 | 5473 | 0.019 | -0.0019 | No | ||
| 66 | ASXL2 | 130670 | 5645 | 0.018 | -0.0110 | No | ||
| 67 | ZCCHC14 | 5900288 | 5811 | 0.016 | -0.0197 | No | ||
| 68 | ARHGAP12 | 1050181 | 5828 | 0.016 | -0.0203 | No | ||
| 69 | STARD13 | 540722 | 5844 | 0.016 | -0.0209 | No | ||
| 70 | FSTL5 | 770280 | 5914 | 0.015 | -0.0245 | No | ||
| 71 | ZFPM2 | 460072 | 5951 | 0.015 | -0.0262 | No | ||
| 72 | SOCS5 | 3830398 7100093 | 5972 | 0.015 | -0.0271 | No | ||
| 73 | PTP4A1 | 130692 | 6117 | 0.014 | -0.0347 | No | ||
| 74 | PLAA | 60706 6370369 | 6186 | 0.013 | -0.0382 | No | ||
| 75 | HIVEP2 | 6520538 | 6207 | 0.013 | -0.0391 | No | ||
| 76 | MAPRE3 | 7050504 | 6232 | 0.013 | -0.0402 | No | ||
| 77 | QKI | 5220093 6130044 | 6276 | 0.013 | -0.0424 | No | ||
| 78 | MLLT6 | 3870168 | 6281 | 0.013 | -0.0424 | No | ||
| 79 | MAP3K9 | 3290707 | 6538 | 0.011 | -0.0562 | No | ||
| 80 | USP33 | 780047 | 6555 | 0.011 | -0.0569 | No | ||
| 81 | AKAP7 | 4610044 | 6695 | 0.010 | -0.0643 | No | ||
| 82 | WDFY3 | 3610041 4560333 4570273 | 6839 | 0.010 | -0.0720 | No | ||
| 83 | BIRC6 | 70148 | 6915 | 0.009 | -0.0759 | No | ||
| 84 | PLCB1 | 2570050 7100102 | 6938 | 0.009 | -0.0770 | No | ||
| 85 | ZBTB4 | 6450441 | 6973 | 0.009 | -0.0787 | No | ||
| 86 | DLL1 | 1770377 | 7009 | 0.009 | -0.0805 | No | ||
| 87 | SULF1 | 430575 | 7131 | 0.008 | -0.0869 | No | ||
| 88 | SOCS6 | 1170347 7100280 | 7409 | 0.007 | -0.1019 | No | ||
| 89 | AKAP1 | 110148 1740735 2260019 7000563 | 7514 | 0.007 | -0.1075 | No | ||
| 90 | RUNX2 | 1400193 4230215 | 7615 | 0.007 | -0.1128 | No | ||
| 91 | WEE1 | 3390070 | 7696 | 0.006 | -0.1171 | No | ||
| 92 | ATRX | 2340039 3610132 4210373 | 7963 | 0.005 | -0.1315 | No | ||
| 93 | ERBB2IP | 580253 1090672 | 8001 | 0.005 | -0.1334 | No | ||
| 94 | PTGFRN | 4120524 | 8221 | 0.005 | -0.1453 | No | ||
| 95 | M6PR | 2480458 3290593 3840324 | 8237 | 0.004 | -0.1461 | No | ||
| 96 | MAMDC1 | 3710129 4480075 | 8251 | 0.004 | -0.1467 | No | ||
| 97 | EMX2 | 1660092 | 8392 | 0.004 | -0.1543 | No | ||
| 98 | NPTX1 | 3710736 | 8394 | 0.004 | -0.1543 | No | ||
| 99 | GAP43 | 3130504 | 8425 | 0.004 | -0.1558 | No | ||
| 100 | RAPGEF4 | 6100563 | 8473 | 0.004 | -0.1583 | No | ||
| 101 | SNX5 | 1570021 6860609 | 8488 | 0.004 | -0.1590 | No | ||
| 102 | CALM2 | 6620463 | 8534 | 0.004 | -0.1614 | No | ||
| 103 | SLC24A3 | 5080707 | 8573 | 0.004 | -0.1635 | No | ||
| 104 | LRCH2 | 2900162 | 8604 | 0.003 | -0.1650 | No | ||
| 105 | ARHGEF12 | 3990195 | 8868 | 0.003 | -0.1793 | No | ||
| 106 | EDG1 | 4200619 | 8898 | 0.003 | -0.1809 | No | ||
| 107 | LDLR | 5670386 | 8928 | 0.002 | -0.1824 | No | ||
| 108 | WNT1 | 4780148 | 8974 | 0.002 | -0.1848 | No | ||
| 109 | FOXF2 | 1240091 | 9092 | 0.002 | -0.1912 | No | ||
| 110 | BTBD7 | 730288 | 9131 | 0.002 | -0.1932 | No | ||
| 111 | CNOT7 | 2450338 5720397 | 9290 | 0.001 | -0.2018 | No | ||
| 112 | NHLH2 | 6200021 | 9445 | 0.001 | -0.2102 | No | ||
| 113 | ADCY2 | 4610100 | 9447 | 0.001 | -0.2102 | No | ||
| 114 | ARHGAP21 | 2100168 | 9474 | 0.001 | -0.2116 | No | ||
| 115 | UBE2D1 | 1850400 | 9493 | 0.001 | -0.2126 | No | ||
| 116 | EXOC5 | 7000711 | 9495 | 0.001 | -0.2126 | No | ||
| 117 | MAP3K12 | 2470373 6420162 | 9831 | -0.000 | -0.2309 | No | ||
| 118 | ST8SIA3 | 6940427 | 9878 | -0.000 | -0.2334 | No | ||
| 119 | ABCA1 | 6290156 | 9882 | -0.000 | -0.2335 | No | ||
| 120 | HOXB3 | 520292 | 9888 | -0.000 | -0.2338 | No | ||
| 121 | NRP2 | 4070400 5860041 6650446 | 9922 | -0.000 | -0.2356 | No | ||
| 122 | UCP3 | 4200128 4210148 | 9927 | -0.000 | -0.2358 | No | ||
| 123 | GDA | 3290435 | 9947 | -0.000 | -0.2368 | No | ||
| 124 | FMR1 | 5050075 | 9995 | -0.001 | -0.2394 | No | ||
| 125 | BHLHB3 | 3450438 | 10001 | -0.001 | -0.2397 | No | ||
| 126 | SHANK2 | 1500452 | 10059 | -0.001 | -0.2428 | No | ||
| 127 | CLCN5 | 4200390 | 10276 | -0.001 | -0.2545 | No | ||
| 128 | ABHD3 | 2370091 | 10318 | -0.002 | -0.2567 | No | ||
| 129 | NPAT | 3800594 4670026 | 10414 | -0.002 | -0.2619 | No | ||
| 130 | MYT1 | 6770524 | 10449 | -0.002 | -0.2637 | No | ||
| 131 | PTPRG | 2650524 1340022 | 10604 | -0.002 | -0.2720 | No | ||
| 132 | TRIM2 | 430546 1170286 | 10606 | -0.002 | -0.2721 | No | ||
| 133 | ROBO1 | 3710398 | 10713 | -0.003 | -0.2778 | No | ||
| 134 | ERBB4 | 4610400 4810017 | 10735 | -0.003 | -0.2789 | No | ||
| 135 | R3HDM1 | 940685 3290161 | 10802 | -0.003 | -0.2825 | No | ||
| 136 | EIF2C1 | 6900551 | 10857 | -0.003 | -0.2854 | No | ||
| 137 | ATP11A | 1240717 5570603 | 10950 | -0.003 | -0.2903 | No | ||
| 138 | TSC1 | 1850672 | 10975 | -0.003 | -0.2916 | No | ||
| 139 | MEOX2 | 6400288 | 11072 | -0.004 | -0.2967 | No | ||
| 140 | NEUROD1 | 3060619 | 11107 | -0.004 | -0.2985 | No | ||
| 141 | EFNB2 | 5340136 | 11171 | -0.004 | -0.3019 | No | ||
| 142 | POU3F2 | 6620484 | 11225 | -0.004 | -0.3047 | No | ||
| 143 | TAF4 | 1450170 | 11332 | -0.005 | -0.3104 | No | ||
| 144 | POU4F1 | 2260315 | 11338 | -0.005 | -0.3107 | No | ||
| 145 | TESK2 | 6520524 | 11378 | -0.005 | -0.3127 | No | ||
| 146 | MAT2B | 1690139 2510706 | 11381 | -0.005 | -0.3127 | No | ||
| 147 | B4GALT5 | 1230692 | 11422 | -0.005 | -0.3149 | No | ||
| 148 | RAI2 | 6770333 | 11595 | -0.006 | -0.3241 | No | ||
| 149 | RXRA | 3800471 | 11617 | -0.006 | -0.3252 | No | ||
| 150 | GRM7 | 4230110 | 11793 | -0.006 | -0.3346 | No | ||
| 151 | RAB5A | 6520324 | 11803 | -0.006 | -0.3350 | No | ||
| 152 | PGM2L1 | 2350035 | 11865 | -0.007 | -0.3383 | No | ||
| 153 | SCRT1 | 6760736 | 11956 | -0.007 | -0.3431 | No | ||
| 154 | ATP2B2 | 3780397 | 12061 | -0.007 | -0.3486 | No | ||
| 155 | DOCK3 | 3290273 3360497 2850068 | 12070 | -0.007 | -0.3489 | No | ||
| 156 | ST18 | 5720537 | 12151 | -0.008 | -0.3532 | No | ||
| 157 | NRBF2 | 6110672 | 12340 | -0.009 | -0.3633 | No | ||
| 158 | MAFB | 1230471 | 12497 | -0.009 | -0.3717 | No | ||
| 159 | PHACTR2 | 1580154 | 12556 | -0.010 | -0.3747 | No | ||
| 160 | TRIM3 | 5570156 | 12623 | -0.010 | -0.3781 | No | ||
| 161 | CREB5 | 2320368 | 12683 | -0.010 | -0.3812 | No | ||
| 162 | WDR20 | 1240373 | 12864 | -0.011 | -0.3908 | No | ||
| 163 | BMPR2 | 5340066 6960328 | 13057 | -0.013 | -0.4011 | No | ||
| 164 | EREG | 50519 4920129 | 13196 | -0.014 | -0.4084 | No | ||
| 165 | HOXA5 | 6840026 | 13239 | -0.014 | -0.4105 | No | ||
| 166 | SOX5 | 2370576 2900167 3190128 5050528 | 13250 | -0.014 | -0.4108 | No | ||
| 167 | CSMD1 | 1780672 | 13316 | -0.015 | -0.4142 | No | ||
| 168 | ADAMTS18 | 5690008 | 13348 | -0.015 | -0.4156 | No | ||
| 169 | WNK3 | 4060053 | 13431 | -0.016 | -0.4199 | No | ||
| 170 | ABCC5 | 2100600 5050692 | 13549 | -0.017 | -0.4260 | No | ||
| 171 | SLC9A2 | 4920022 5910133 | 13559 | -0.017 | -0.4262 | No | ||
| 172 | BTBD3 | 1980041 2510577 3190053 4200746 | 13823 | -0.020 | -0.4403 | No | ||
| 173 | NPNT | 130301 | 13826 | -0.020 | -0.4401 | No | ||
| 174 | COL19A1 | 6220441 | 13920 | -0.022 | -0.4448 | No | ||
| 175 | ATRN | 870735 3360390 | 13936 | -0.022 | -0.4453 | No | ||
| 176 | TARDBP | 870390 2350093 | 13953 | -0.022 | -0.4458 | No | ||
| 177 | PAFAH1B1 | 4230333 6420121 6450066 | 14002 | -0.023 | -0.4481 | No | ||
| 178 | LNPEP | 1170092 1940093 3120025 | 14085 | -0.024 | -0.4522 | No | ||
| 179 | ADAM12 | 3390132 4070347 | 14089 | -0.024 | -0.4520 | No | ||
| 180 | DCBLD2 | 5890333 | 14090 | -0.024 | -0.4517 | No | ||
| 181 | NIPA1 | 380600 | 14166 | -0.026 | -0.4554 | No | ||
| 182 | LRP8 | 3610746 5360035 | 14178 | -0.026 | -0.4556 | No | ||
| 183 | ZNF3 | 1770193 2320047 2570273 | 14212 | -0.026 | -0.4570 | No | ||
| 184 | WNT2B | 3130088 5960575 | 14368 | -0.029 | -0.4650 | No | ||
| 185 | TBC1D8 | 2340253 | 14517 | -0.032 | -0.4726 | No | ||
| 186 | DPYSL2 | 2100427 3130112 5700324 | 14586 | -0.035 | -0.4758 | No | ||
| 187 | TMEPAI | 1340253 | 14599 | -0.035 | -0.4759 | No | ||
| 188 | EIF2C4 | 2940577 5900563 | 14615 | -0.035 | -0.4762 | No | ||
| 189 | ARHGAP1 | 2810010 5270064 | 14636 | -0.036 | -0.4768 | No | ||
| 190 | CSNK1G1 | 840082 1230575 3940647 | 14719 | -0.039 | -0.4807 | No | ||
| 191 | CNOT4 | 2320242 6760706 | 14751 | -0.040 | -0.4818 | No | ||
| 192 | CHD9 | 430037 2570129 5420398 | 15012 | -0.053 | -0.4952 | No | ||
| 193 | SNF1LK | 6110403 | 15238 | -0.067 | -0.5064 | No | ||
| 194 | SPATA2 | 1230215 5910541 | 15316 | -0.073 | -0.5096 | No | ||
| 195 | MLLT10 | 1410288 3290142 | 15358 | -0.077 | -0.5107 | No | ||
| 196 | PIGA | 1940435 5290692 | 15419 | -0.082 | -0.5127 | No | ||
| 197 | PAPD4 | 1580673 730605 5270112 | 15515 | -0.090 | -0.5166 | No | ||
| 198 | CLOCK | 1410070 2940292 3390075 | 15517 | -0.090 | -0.5153 | No | ||
| 199 | SUV39H1 | 630112 | 15532 | -0.092 | -0.5147 | No | ||
| 200 | NEUROG1 | 380341 | 15544 | -0.094 | -0.5139 | No | ||
| 201 | VGLL4 | 6860463 | 15605 | -0.101 | -0.5157 | No | ||
| 202 | E2F7 | 4200056 | 15660 | -0.106 | -0.5171 | No | ||
| 203 | MTF1 | 520427 2340364 | 15674 | -0.108 | -0.5162 | No | ||
| 204 | TIMP2 | 4780609 | 15713 | -0.111 | -0.5166 | No | ||
| 205 | RNF38 | 1940746 3140133 3840601 | 15724 | -0.113 | -0.5155 | No | ||
| 206 | STX6 | 4850750 6770278 | 15798 | -0.120 | -0.5177 | No | ||
| 207 | BRWD1 | 3130048 3140168 6400601 | 15850 | -0.126 | -0.5186 | No | ||
| 208 | DICER1 | 540286 | 15855 | -0.127 | -0.5170 | No | ||
| 209 | ACBD5 | 3440397 6020600 | 15906 | -0.131 | -0.5178 | No | ||
| 210 | USP48 | 2480010 | 16050 | -0.149 | -0.5234 | No | ||
| 211 | AP1G1 | 2360671 | 16058 | -0.151 | -0.5215 | No | ||
| 212 | CCRN4L | 50438 5130372 | 16181 | -0.169 | -0.5257 | No | ||
| 213 | RKHD1 | 6860577 | 16222 | -0.174 | -0.5253 | No | ||
| 214 | RAB5B | 130397 1980273 6900731 | 16259 | -0.179 | -0.5246 | No | ||
| 215 | ATXN1 | 5550156 | 16338 | -0.192 | -0.5260 | No | ||
| 216 | SNIP1 | 5360092 | 16535 | -0.233 | -0.5333 | No | ||
| 217 | SMARCD2 | 3830619 | 16579 | -0.244 | -0.5320 | No | ||
| 218 | DNM2 | 50176 3170279 | 16680 | -0.265 | -0.5335 | No | ||
| 219 | BZRAP1 | 5290487 | 16725 | -0.275 | -0.5319 | No | ||
| 220 | ULK2 | 50041 5050176 | 16965 | -0.339 | -0.5399 | Yes | ||
| 221 | VPS24 | 3120687 | 17035 | -0.357 | -0.5384 | Yes | ||
| 222 | EPS15 | 4200215 | 17038 | -0.358 | -0.5332 | Yes | ||
| 223 | MECP2 | 1940450 | 17044 | -0.360 | -0.5282 | Yes | ||
| 224 | PFKFB3 | 630706 | 17072 | -0.370 | -0.5242 | Yes | ||
| 225 | TNRC6A | 1240537 6130037 6200010 | 17167 | -0.403 | -0.5234 | Yes | ||
| 226 | OGT | 2360131 4610333 | 17176 | -0.406 | -0.5178 | Yes | ||
| 227 | HECA | 3310612 | 17186 | -0.409 | -0.5123 | Yes | ||
| 228 | ENPP5 | 1780338 | 17203 | -0.414 | -0.5071 | Yes | ||
| 229 | RALBP1 | 4780632 | 17268 | -0.439 | -0.5041 | Yes | ||
| 230 | MTMR12 | 7000181 | 17366 | -0.480 | -0.5023 | Yes | ||
| 231 | PHF12 | 870400 3130687 | 17447 | -0.516 | -0.4990 | Yes | ||
| 232 | MYB | 1660494 5860451 6130706 | 17450 | -0.518 | -0.4915 | Yes | ||
| 233 | HRB | 1660397 6110398 | 17455 | -0.520 | -0.4840 | Yes | ||
| 234 | ITPK1 | 3360056 | 17460 | -0.522 | -0.4766 | Yes | ||
| 235 | ATG16L1 | 5390292 | 17489 | -0.535 | -0.4702 | Yes | ||
| 236 | MAP3K14 | 5890435 | 17492 | -0.536 | -0.4624 | Yes | ||
| 237 | BAZ2A | 730184 | 17621 | -0.608 | -0.4604 | Yes | ||
| 238 | ITPR1 | 3450519 | 17658 | -0.631 | -0.4531 | Yes | ||
| 239 | SOX4 | 2260091 | 17671 | -0.637 | -0.4443 | Yes | ||
| 240 | IRF1 | 2340152 3450592 6290121 6980577 | 17795 | -0.709 | -0.4406 | Yes | ||
| 241 | NCOA1 | 3610438 | 17802 | -0.715 | -0.4304 | Yes | ||
| 242 | NDEL1 | 2370465 | 17853 | -0.750 | -0.4220 | Yes | ||
| 243 | BACH2 | 6760131 | 17928 | -0.809 | -0.4141 | Yes | ||
| 244 | ATP6V1B2 | 4060528 | 17939 | -0.818 | -0.4026 | Yes | ||
| 245 | PELI1 | 3870215 6900040 | 17968 | -0.838 | -0.3918 | Yes | ||
| 246 | SPEN | 2060041 | 18077 | -0.965 | -0.3834 | Yes | ||
| 247 | GADD45A | 2900717 | 18078 | -0.966 | -0.3692 | Yes | ||
| 248 | PSAP | 2810338 | 18107 | -0.996 | -0.3560 | Yes | ||
| 249 | ZFYVE26 | 6860152 | 18188 | -1.106 | -0.3441 | Yes | ||
| 250 | KLF13 | 4670563 | 18251 | -1.209 | -0.3296 | Yes | ||
| 251 | PITPNM2 | 1580044 | 18258 | -1.232 | -0.3118 | Yes | ||
| 252 | MAML1 | 2760008 | 18267 | -1.263 | -0.2936 | Yes | ||
| 253 | DDX6 | 2630025 6770408 | 18287 | -1.309 | -0.2754 | Yes | ||
| 254 | WDR1 | 1090129 3990121 | 18353 | -1.479 | -0.2571 | Yes | ||
| 255 | SPHK2 | 2030487 4120670 4850114 | 18441 | -1.776 | -0.2356 | Yes | ||
| 256 | HBP1 | 2510102 3130010 4210619 | 18447 | -1.783 | -0.2096 | Yes | ||
| 257 | LRIG1 | 1050136 | 18478 | -1.980 | -0.1821 | Yes | ||
| 258 | CNIH | 2190484 5080609 | 18545 | -2.546 | -0.1481 | Yes | ||
| 259 | ELK3 | 460377 1400762 3710497 | 18561 | -2.759 | -0.1083 | Yes | ||
| 260 | SMOC1 | 840039 | 18589 | -3.340 | -0.0605 | Yes | ||
| 261 | BTG1 | 4200735 6040131 6200133 | 18607 | -4.199 | 0.0005 | Yes |

