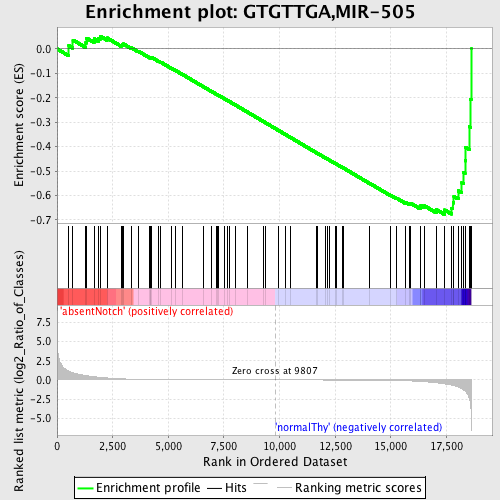
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GTGTTGA,MIR-505 |
| Enrichment Score (ES) | -0.67757344 |
| Normalized Enrichment Score (NES) | -1.5316963 |
| Nominal p-value | 0.0046948357 |
| FDR q-value | 0.17100127 |
| FWER p-Value | 0.644 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDX3X | 2190020 | 528 | 1.119 | 0.0127 | No | ||
| 2 | MBNL1 | 2640762 7100048 | 713 | 0.914 | 0.0364 | No | ||
| 3 | ALS2 | 3130546 6400070 | 1293 | 0.561 | 0.0258 | No | ||
| 4 | SLC25A3 | 670112 | 1327 | 0.546 | 0.0441 | No | ||
| 5 | SIRT3 | 4010440 6040431 | 1664 | 0.415 | 0.0413 | No | ||
| 6 | PMP22 | 4010239 6550072 | 1862 | 0.356 | 0.0438 | No | ||
| 7 | DHX15 | 870632 | 1932 | 0.335 | 0.0524 | No | ||
| 8 | KRAS | 2060170 | 2245 | 0.255 | 0.0449 | No | ||
| 9 | CAMK2D | 2370685 | 2879 | 0.142 | 0.0160 | No | ||
| 10 | YTHDF3 | 4540632 4670717 7040609 | 2938 | 0.134 | 0.0178 | No | ||
| 11 | UBE2D3 | 3190452 | 2980 | 0.131 | 0.0204 | No | ||
| 12 | CITED2 | 5670114 5130088 | 3340 | 0.095 | 0.0046 | No | ||
| 13 | STRN3 | 1450093 6200685 | 3641 | 0.073 | -0.0089 | No | ||
| 14 | CTDSPL | 4670546 | 4168 | 0.047 | -0.0355 | No | ||
| 15 | GLP1R | 6420528 | 4210 | 0.046 | -0.0361 | No | ||
| 16 | EPN2 | 1340451 6550239 6860020 6860403 | 4213 | 0.045 | -0.0345 | No | ||
| 17 | CANX | 4760402 | 4220 | 0.045 | -0.0332 | No | ||
| 18 | PDE4D | 2470528 6660014 | 4558 | 0.035 | -0.0501 | No | ||
| 19 | RTF1 | 2100735 | 4664 | 0.032 | -0.0545 | No | ||
| 20 | MXI1 | 5050064 5130484 | 5146 | 0.024 | -0.0796 | No | ||
| 21 | USP47 | 2640497 | 5322 | 0.021 | -0.0883 | No | ||
| 22 | SFRS1 | 2360440 | 5619 | 0.018 | -0.1035 | No | ||
| 23 | KCNJ2 | 630019 | 6578 | 0.011 | -0.1548 | No | ||
| 24 | EIF3S10 | 1190079 3840176 | 6955 | 0.009 | -0.1747 | No | ||
| 25 | LIN28 | 6590672 | 7157 | 0.008 | -0.1853 | No | ||
| 26 | HNT | 6450731 | 7215 | 0.008 | -0.1880 | No | ||
| 27 | FIGN | 7000601 | 7259 | 0.008 | -0.1901 | No | ||
| 28 | AKAP1 | 110148 1740735 2260019 7000563 | 7514 | 0.007 | -0.2035 | No | ||
| 29 | SRGAP2 | 1050092 1230403 3830066 6130215 | 7643 | 0.007 | -0.2102 | No | ||
| 30 | PCDH9 | 2100136 | 7761 | 0.006 | -0.2162 | No | ||
| 31 | GCC1 | 2510132 | 8008 | 0.005 | -0.2293 | No | ||
| 32 | CREB1 | 1500717 2230358 3610600 6550601 | 8539 | 0.004 | -0.2577 | No | ||
| 33 | ZIC2 | 1410408 | 9255 | 0.002 | -0.2962 | No | ||
| 34 | SLC18A2 | 6420541 | 9359 | 0.001 | -0.3017 | No | ||
| 35 | GDA | 3290435 | 9947 | -0.000 | -0.3334 | No | ||
| 36 | ZMYND11 | 630181 2570019 4850138 5360040 | 10251 | -0.001 | -0.3497 | No | ||
| 37 | DPYSL3 | 1580594 | 10478 | -0.002 | -0.3618 | No | ||
| 38 | CUGBP1 | 450292 510022 7050176 7050215 | 11641 | -0.006 | -0.4242 | No | ||
| 39 | SON | 3120148 5860239 7040403 | 11712 | -0.006 | -0.4278 | No | ||
| 40 | ATP2B2 | 3780397 | 12061 | -0.007 | -0.4463 | No | ||
| 41 | FZD8 | 1990053 | 12131 | -0.008 | -0.4497 | No | ||
| 42 | MLLT7 | 4480707 | 12258 | -0.008 | -0.4562 | No | ||
| 43 | MAFB | 1230471 | 12497 | -0.009 | -0.4687 | No | ||
| 44 | ELAVL2 | 360181 | 12551 | -0.010 | -0.4712 | No | ||
| 45 | CYP46A1 | 2470332 | 12840 | -0.011 | -0.4863 | No | ||
| 46 | DDX3Y | 1580278 4200519 | 12883 | -0.012 | -0.4881 | No | ||
| 47 | GPR85 | 5130750 5910020 | 14047 | -0.024 | -0.5499 | No | ||
| 48 | AKAP2 | 5550347 | 15004 | -0.052 | -0.5996 | No | ||
| 49 | KHDRBS1 | 1240403 6040040 | 15268 | -0.070 | -0.6112 | No | ||
| 50 | B4GALT1 | 6980167 | 15642 | -0.104 | -0.6275 | No | ||
| 51 | HIPK1 | 110193 | 15832 | -0.125 | -0.6331 | No | ||
| 52 | RANBP9 | 4670685 | 15898 | -0.130 | -0.6318 | No | ||
| 53 | BICD2 | 1090411 | 16324 | -0.190 | -0.6477 | No | ||
| 54 | SLC12A2 | 4610019 6040672 | 16349 | -0.195 | -0.6418 | No | ||
| 55 | MAPK14 | 5290731 | 16518 | -0.228 | -0.6425 | No | ||
| 56 | MECP2 | 1940450 | 17044 | -0.360 | -0.6575 | No | ||
| 57 | MKRN1 | 1410097 | 17417 | -0.502 | -0.6591 | Yes | ||
| 58 | CGGBP1 | 4810053 | 17727 | -0.671 | -0.6511 | Yes | ||
| 59 | NFE2L1 | 6130053 | 17794 | -0.709 | -0.6285 | Yes | ||
| 60 | RUNX1 | 3840711 | 17835 | -0.736 | -0.6036 | Yes | ||
| 61 | H2AFV | 2030577 5220129 | 18032 | -0.904 | -0.5809 | Yes | ||
| 62 | RNF44 | 5910692 | 18193 | -1.116 | -0.5485 | Yes | ||
| 63 | PDPK1 | 6650168 | 18263 | -1.250 | -0.5062 | Yes | ||
| 64 | BCL11B | 2680673 | 18339 | -1.434 | -0.4574 | Yes | ||
| 65 | ARNTL | 3170463 | 18364 | -1.517 | -0.4029 | Yes | ||
| 66 | CNIH | 2190484 5080609 | 18545 | -2.546 | -0.3189 | Yes | ||
| 67 | CYFIP2 | 520577 6040300 | 18580 | -3.091 | -0.2070 | Yes | ||
| 68 | HRBL | 1850102 3830253 4780176 | 18611 | -5.676 | 0.0003 | Yes |

