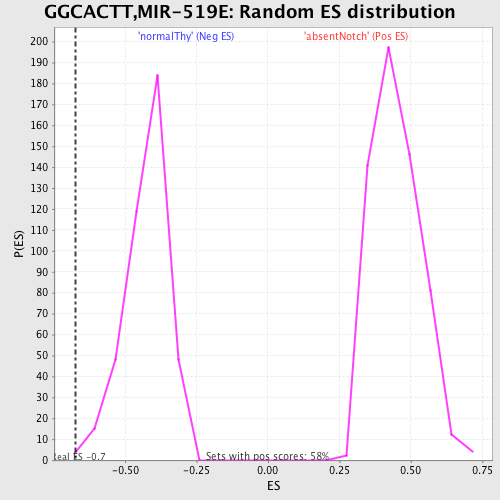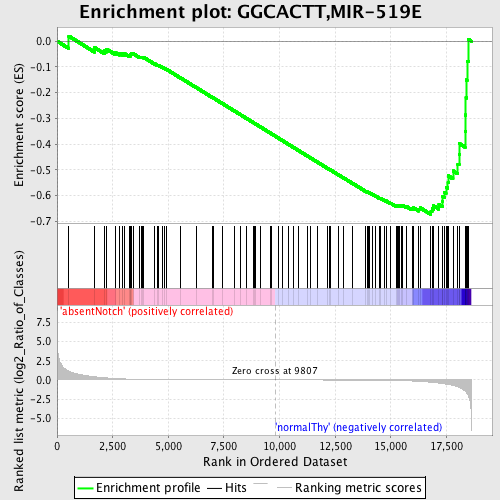
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | GGCACTT,MIR-519E |
| Enrichment Score (ES) | -0.6732477 |
| Normalized Enrichment Score (NES) | -1.5914805 |
| Nominal p-value | 0.004796163 |
| FDR q-value | 0.20786686 |
| FWER p-Value | 0.361 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DDX3X | 2190020 | 528 | 1.119 | 0.0197 | No | ||
| 2 | ZNF148 | 6420403 | 1677 | 0.410 | -0.0246 | No | ||
| 3 | DAZAP2 | 6200102 | 2141 | 0.281 | -0.0375 | No | ||
| 4 | RPS6KA3 | 1980707 | 2228 | 0.258 | -0.0310 | No | ||
| 5 | MAP3K7 | 6040068 | 2614 | 0.182 | -0.0439 | No | ||
| 6 | ZBTB7A | 3780092 | 2800 | 0.153 | -0.0473 | No | ||
| 7 | SEMA4G | 5890026 | 2928 | 0.135 | -0.0484 | No | ||
| 8 | C1GALT1 | 6550088 | 3039 | 0.124 | -0.0489 | No | ||
| 9 | PPP2R2A | 2900014 5700575 | 3231 | 0.104 | -0.0548 | No | ||
| 10 | NR4A2 | 60273 | 3289 | 0.098 | -0.0536 | No | ||
| 11 | MMD | 3850239 | 3313 | 0.097 | -0.0507 | No | ||
| 12 | MYST3 | 5270500 | 3341 | 0.094 | -0.0481 | No | ||
| 13 | MMP19 | 5080377 | 3422 | 0.088 | -0.0486 | No | ||
| 14 | FGD1 | 4780021 | 3721 | 0.069 | -0.0617 | No | ||
| 15 | ARID4B | 730754 6660026 | 3777 | 0.065 | -0.0619 | No | ||
| 16 | TBL1X | 6400524 | 3820 | 0.062 | -0.0615 | No | ||
| 17 | NPAS2 | 770670 | 3895 | 0.059 | -0.0629 | No | ||
| 18 | NR2F2 | 3170609 3310577 | 4398 | 0.040 | -0.0883 | No | ||
| 19 | SOX11 | 610279 | 4515 | 0.036 | -0.0930 | No | ||
| 20 | ST8SIA2 | 2680082 | 4545 | 0.035 | -0.0931 | No | ||
| 21 | VPS26A | 1940494 5720059 | 4731 | 0.031 | -0.1017 | No | ||
| 22 | DPYSL5 | 4900333 | 4814 | 0.029 | -0.1049 | No | ||
| 23 | MTMR4 | 4540524 | 4916 | 0.027 | -0.1092 | No | ||
| 24 | LBP | 6860019 | 5567 | 0.018 | -0.1435 | No | ||
| 25 | QKI | 5220093 6130044 | 6276 | 0.013 | -0.1812 | No | ||
| 26 | ZBTB4 | 6450441 | 6973 | 0.009 | -0.2184 | No | ||
| 27 | DLL1 | 1770377 | 7009 | 0.009 | -0.2199 | No | ||
| 28 | BCOR | 1660452 3940053 | 7426 | 0.007 | -0.2420 | No | ||
| 29 | SERTAD2 | 2810673 6840450 | 7973 | 0.005 | -0.2713 | No | ||
| 30 | PTGFRN | 4120524 | 8221 | 0.005 | -0.2844 | No | ||
| 31 | LPHN2 | 4480010 | 8519 | 0.004 | -0.3003 | No | ||
| 32 | PAFAH1B2 | 5360546 6860672 | 8816 | 0.003 | -0.3161 | No | ||
| 33 | ARHGEF12 | 3990195 | 8868 | 0.003 | -0.3188 | No | ||
| 34 | EDG1 | 4200619 | 8898 | 0.003 | -0.3202 | No | ||
| 35 | BTBD7 | 730288 | 9131 | 0.002 | -0.3327 | No | ||
| 36 | CCNT2 | 2100390 4150563 | 9605 | 0.001 | -0.3582 | No | ||
| 37 | WAC | 5340390 5910164 | 9643 | 0.001 | -0.3601 | No | ||
| 38 | PCYT1B | 1780402 | 9970 | -0.001 | -0.3777 | No | ||
| 39 | MYLK | 4010600 7000364 | 10133 | -0.001 | -0.3864 | No | ||
| 40 | PTPN3 | 870369 | 10412 | -0.002 | -0.4014 | No | ||
| 41 | PTPRG | 2650524 1340022 | 10604 | -0.002 | -0.4116 | No | ||
| 42 | EIF2C1 | 6900551 | 10857 | -0.003 | -0.4250 | No | ||
| 43 | EIF5A2 | 7040139 | 11261 | -0.004 | -0.4466 | No | ||
| 44 | MAT2B | 1690139 2510706 | 11381 | -0.005 | -0.4528 | No | ||
| 45 | SON | 3120148 5860239 7040403 | 11712 | -0.006 | -0.4704 | No | ||
| 46 | SCML2 | 3290603 | 12153 | -0.008 | -0.4938 | No | ||
| 47 | MGEA5 | 6100014 | 12244 | -0.008 | -0.4983 | No | ||
| 48 | CORO7 | 6940309 | 12282 | -0.008 | -0.4999 | No | ||
| 49 | EFNB3 | 5570594 | 12635 | -0.010 | -0.5185 | No | ||
| 50 | DDX3Y | 1580278 4200519 | 12883 | -0.012 | -0.5313 | No | ||
| 51 | DKK1 | 1940215 | 13279 | -0.014 | -0.5520 | No | ||
| 52 | ZFHX4 | 6110167 | 13865 | -0.021 | -0.5827 | No | ||
| 53 | SSFA2 | 3870133 | 13931 | -0.022 | -0.5853 | No | ||
| 54 | PAFAH1B1 | 4230333 6420121 6450066 | 14002 | -0.023 | -0.5881 | No | ||
| 55 | EPHA7 | 1190288 4010278 | 14056 | -0.024 | -0.5899 | No | ||
| 56 | ARID4A | 520156 1050707 | 14162 | -0.026 | -0.5945 | No | ||
| 57 | TAGAP | 3290433 | 14320 | -0.028 | -0.6018 | No | ||
| 58 | PTPRD | 520008 3120097 | 14507 | -0.032 | -0.6104 | No | ||
| 59 | AOF1 | 60121 540184 1340315 | 14524 | -0.033 | -0.6099 | No | ||
| 60 | NANOS1 | 1300402 | 14709 | -0.038 | -0.6181 | No | ||
| 61 | FURIN | 4120168 | 14787 | -0.042 | -0.6205 | No | ||
| 62 | SP1 | 6590017 | 14989 | -0.051 | -0.6291 | No | ||
| 63 | PHF1 | 4010577 6420053 | 15233 | -0.067 | -0.6394 | No | ||
| 64 | SPATA2 | 1230215 5910541 | 15316 | -0.073 | -0.6407 | No | ||
| 65 | XRN1 | 5900093 | 15334 | -0.074 | -0.6384 | No | ||
| 66 | ZNF385 | 450176 4810358 | 15405 | -0.080 | -0.6387 | No | ||
| 67 | CDK10 | 60164 1190717 3130128 3990044 | 15484 | -0.087 | -0.6392 | No | ||
| 68 | CLOCK | 1410070 2940292 3390075 | 15517 | -0.090 | -0.6370 | No | ||
| 69 | MARK4 | 730519 | 15698 | -0.110 | -0.6420 | No | ||
| 70 | OSR1 | 1500025 | 15952 | -0.137 | -0.6497 | No | ||
| 71 | ZIC4 | 1500082 | 16025 | -0.146 | -0.6473 | No | ||
| 72 | RAB5B | 130397 1980273 6900731 | 16259 | -0.179 | -0.6522 | No | ||
| 73 | BICD2 | 1090411 | 16324 | -0.190 | -0.6474 | No | ||
| 74 | PERQ1 | 1340164 | 16803 | -0.295 | -0.6605 | Yes | ||
| 75 | RRM2 | 6350059 6940162 | 16853 | -0.307 | -0.6500 | Yes | ||
| 76 | FBXO21 | 6550070 | 16921 | -0.326 | -0.6395 | Yes | ||
| 77 | PTEN | 3390064 | 17161 | -0.403 | -0.6351 | Yes | ||
| 78 | CXCR4 | 4590519 | 17323 | -0.464 | -0.6238 | Yes | ||
| 79 | CCNG2 | 3190095 | 17333 | -0.468 | -0.6041 | Yes | ||
| 80 | MKRN1 | 1410097 | 17417 | -0.502 | -0.5870 | Yes | ||
| 81 | ARHGEF5 | 3170605 | 17483 | -0.534 | -0.5675 | Yes | ||
| 82 | RAB35 | 630056 | 17569 | -0.573 | -0.5474 | Yes | ||
| 83 | SRPK1 | 450110 | 17581 | -0.581 | -0.5230 | Yes | ||
| 84 | IRF1 | 2340152 3450592 6290121 6980577 | 17795 | -0.709 | -0.5039 | Yes | ||
| 85 | MARK2 | 7210608 | 18013 | -0.886 | -0.4775 | Yes | ||
| 86 | NEDD4L | 6380368 | 18070 | -0.959 | -0.4392 | Yes | ||
| 87 | UBE3C | 3140673 | 18089 | -0.976 | -0.3981 | Yes | ||
| 88 | BCL11B | 2680673 | 18339 | -1.434 | -0.3498 | Yes | ||
| 89 | PLAGL2 | 1770148 | 18356 | -1.489 | -0.2865 | Yes | ||
| 90 | ARL4C | 1190279 | 18378 | -1.566 | -0.2202 | Yes | ||
| 91 | EHD3 | 5360484 | 18394 | -1.640 | -0.1504 | Yes | ||
| 92 | RBL2 | 580446 1400670 | 18429 | -1.740 | -0.0772 | Yes | ||
| 93 | BNIP2 | 1410475 6770088 | 18487 | -2.026 | 0.0070 | Yes |