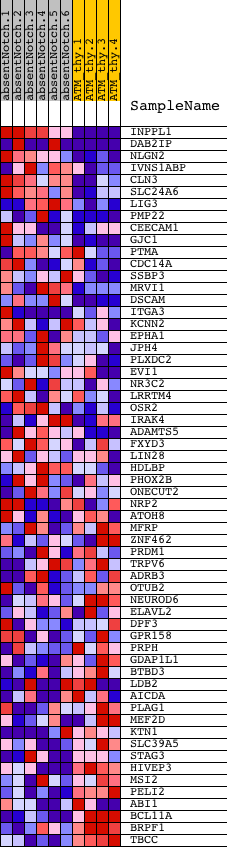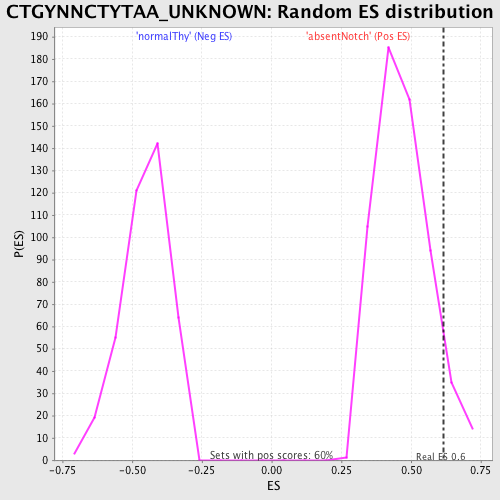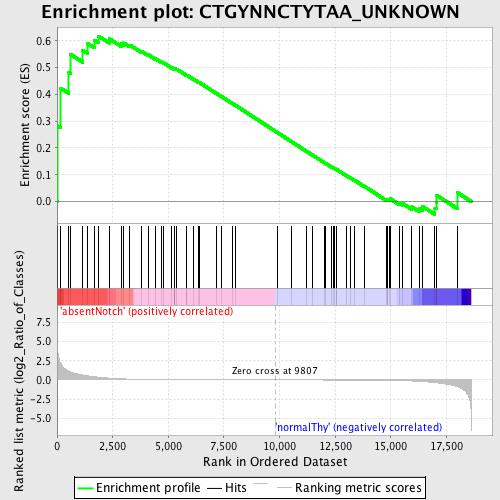
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | CTGYNNCTYTAA_UNKNOWN |
| Enrichment Score (ES) | 0.61733943 |
| Normalized Enrichment Score (NES) | 1.3109906 |
| Nominal p-value | 0.07214765 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | INPPL1 | 110717 3120164 | 27 | 4.033 | 0.2822 | Yes | ||
| 2 | DAB2IP | 3170064 6200441 | 166 | 2.104 | 0.4228 | Yes | ||
| 3 | NLGN2 | 5900592 | 530 | 1.115 | 0.4816 | Yes | ||
| 4 | IVNS1ABP | 4760601 6520113 | 615 | 1.022 | 0.5489 | Yes | ||
| 5 | CLN3 | 780368 | 1147 | 0.635 | 0.5650 | Yes | ||
| 6 | SLC24A6 | 4230068 | 1368 | 0.524 | 0.5900 | Yes | ||
| 7 | LIG3 | 2940300 | 1680 | 0.410 | 0.6021 | Yes | ||
| 8 | PMP22 | 4010239 6550072 | 1862 | 0.356 | 0.6173 | Yes | ||
| 9 | CEECAM1 | 780605 | 2343 | 0.234 | 0.6079 | No | ||
| 10 | GJC1 | 7050082 | 2880 | 0.142 | 0.5890 | No | ||
| 11 | PTMA | 5570148 | 2997 | 0.128 | 0.5918 | No | ||
| 12 | CDC14A | 4050132 | 3274 | 0.100 | 0.5839 | No | ||
| 13 | SSBP3 | 870717 1050377 7040551 | 3782 | 0.065 | 0.5611 | No | ||
| 14 | MRVI1 | 4810338 4850601 5900441 | 4090 | 0.050 | 0.5481 | No | ||
| 15 | DSCAM | 1780050 2450731 2810438 | 4405 | 0.039 | 0.5340 | No | ||
| 16 | ITGA3 | 4570427 | 4687 | 0.032 | 0.5210 | No | ||
| 17 | KCNN2 | 990025 3520524 | 4792 | 0.029 | 0.5175 | No | ||
| 18 | EPHA1 | 4590021 5670139 6660100 | 5156 | 0.023 | 0.4996 | No | ||
| 19 | JPH4 | 780020 | 5261 | 0.022 | 0.4955 | No | ||
| 20 | PLXDC2 | 6400358 | 5281 | 0.022 | 0.4960 | No | ||
| 21 | EVI1 | 2000193 2680494 | 5282 | 0.022 | 0.4975 | No | ||
| 22 | NR3C2 | 2480324 6400093 | 5377 | 0.020 | 0.4939 | No | ||
| 23 | LRRTM4 | 5910100 | 5804 | 0.016 | 0.4721 | No | ||
| 24 | OSR2 | 70711 | 5825 | 0.016 | 0.4721 | No | ||
| 25 | IRAK4 | 5390156 6130594 | 6143 | 0.014 | 0.4560 | No | ||
| 26 | ADAMTS5 | 5890592 | 6349 | 0.012 | 0.4458 | No | ||
| 27 | FXYD3 | 2320450 | 6384 | 0.012 | 0.4449 | No | ||
| 28 | LIN28 | 6590672 | 7157 | 0.008 | 0.4039 | No | ||
| 29 | HDLBP | 2350692 6590520 | 7378 | 0.007 | 0.3925 | No | ||
| 30 | PHOX2B | 5270075 | 7890 | 0.006 | 0.3654 | No | ||
| 31 | ONECUT2 | 1340008 | 8002 | 0.005 | 0.3598 | No | ||
| 32 | NRP2 | 4070400 5860041 6650446 | 9922 | -0.000 | 0.2564 | No | ||
| 33 | ATOH8 | 4540204 | 10526 | -0.002 | 0.2240 | No | ||
| 34 | MFRP | 670541 | 11208 | -0.004 | 0.1876 | No | ||
| 35 | ZNF462 | 6900100 | 11464 | -0.005 | 0.1742 | No | ||
| 36 | PRDM1 | 3170347 3520301 | 11997 | -0.007 | 0.1461 | No | ||
| 37 | TRPV6 | 4010717 | 12068 | -0.007 | 0.1428 | No | ||
| 38 | ADRB3 | 6900072 | 12345 | -0.009 | 0.1286 | No | ||
| 39 | OTUB2 | 2570725 2690647 | 12430 | -0.009 | 0.1247 | No | ||
| 40 | NEUROD6 | 4670731 | 12460 | -0.009 | 0.1238 | No | ||
| 41 | ELAVL2 | 360181 | 12551 | -0.010 | 0.1196 | No | ||
| 42 | DPF3 | 2650128 2970168 | 12571 | -0.010 | 0.1193 | No | ||
| 43 | GPR158 | 4920598 | 13017 | -0.012 | 0.0962 | No | ||
| 44 | PRPH | 2060136 | 13187 | -0.014 | 0.0880 | No | ||
| 45 | GDAP1L1 | 1660066 | 13374 | -0.015 | 0.0791 | No | ||
| 46 | BTBD3 | 1980041 2510577 3190053 4200746 | 13823 | -0.020 | 0.0563 | No | ||
| 47 | LDB2 | 5670441 6110170 | 14784 | -0.041 | 0.0075 | No | ||
| 48 | AICDA | 2340487 3800672 | 14855 | -0.044 | 0.0069 | No | ||
| 49 | PLAG1 | 1450142 3870139 6520039 | 14865 | -0.045 | 0.0095 | No | ||
| 50 | MEF2D | 5690576 | 14956 | -0.050 | 0.0082 | No | ||
| 51 | KTN1 | 70446 3450609 4560048 | 14971 | -0.050 | 0.0109 | No | ||
| 52 | SLC39A5 | 2900100 4610440 | 15389 | -0.079 | -0.0060 | No | ||
| 53 | STAG3 | 3830112 | 15522 | -0.091 | -0.0067 | No | ||
| 54 | HIVEP3 | 3990551 | 15947 | -0.136 | -0.0200 | No | ||
| 55 | MSI2 | 1190066 5270039 | 16271 | -0.180 | -0.0247 | No | ||
| 56 | PELI2 | 1940364 | 16432 | -0.210 | -0.0185 | No | ||
| 57 | ABI1 | 2850152 | 16983 | -0.344 | -0.0240 | No | ||
| 58 | BCL11A | 6860369 | 17052 | -0.364 | -0.0020 | No | ||
| 59 | BRPF1 | 3170139 | 17066 | -0.370 | 0.0233 | No | ||
| 60 | TBCC | 4810021 | 17983 | -0.856 | 0.0341 | No |