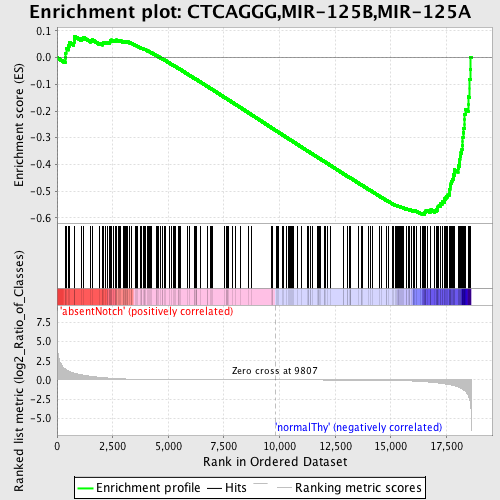
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CTCAGGG,MIR-125B,MIR-125A |
| Enrichment Score (ES) | -0.5874645 |
| Normalized Enrichment Score (NES) | -1.5577937 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2623999 |
| FWER p-Value | 0.514 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC25A15 | 4640368 5550064 | 361 | 1.411 | -0.0004 | No | ||
| 2 | EMID1 | 3190079 5130152 | 385 | 1.364 | 0.0170 | No | ||
| 3 | VANGL2 | 70097 870075 | 400 | 1.345 | 0.0346 | No | ||
| 4 | PPAT | 1980019 | 515 | 1.138 | 0.0439 | No | ||
| 5 | SOCS4 | 1170162 | 540 | 1.107 | 0.0577 | No | ||
| 6 | ATP5G2 | 840068 | 759 | 0.874 | 0.0578 | No | ||
| 7 | NEU1 | 380546 | 765 | 0.870 | 0.0694 | No | ||
| 8 | EIF2C2 | 770451 | 778 | 0.857 | 0.0804 | No | ||
| 9 | BMF | 610102 | 1094 | 0.658 | 0.0722 | No | ||
| 10 | FUT4 | 2030324 | 1175 | 0.618 | 0.0763 | No | ||
| 11 | BAP1 | 3830131 | 1511 | 0.471 | 0.0645 | No | ||
| 12 | RPS6KA1 | 4120647 | 1583 | 0.443 | 0.0667 | No | ||
| 13 | PCGF6 | 6420487 | 1911 | 0.342 | 0.0536 | No | ||
| 14 | UBE2L3 | 1690707 5900242 | 2032 | 0.311 | 0.0513 | No | ||
| 15 | GRSF1 | 2100184 | 2059 | 0.303 | 0.0540 | No | ||
| 16 | ITGA9 | 670180 | 2096 | 0.290 | 0.0560 | No | ||
| 17 | NUP210 | 3610161 | 2160 | 0.275 | 0.0563 | No | ||
| 18 | LOXL1 | 3520537 | 2244 | 0.255 | 0.0553 | No | ||
| 19 | APOBEC1 | 4610022 | 2265 | 0.251 | 0.0576 | No | ||
| 20 | MADD | 6100725 7000088 | 2351 | 0.232 | 0.0561 | No | ||
| 21 | LACTB | 2320139 | 2371 | 0.228 | 0.0582 | No | ||
| 22 | ELOVL6 | 5340746 | 2378 | 0.226 | 0.0610 | No | ||
| 23 | DAG1 | 460053 610341 | 2391 | 0.223 | 0.0634 | No | ||
| 24 | MLF2 | 1940176 | 2423 | 0.219 | 0.0647 | No | ||
| 25 | TRPS1 | 870100 | 2453 | 0.213 | 0.0660 | No | ||
| 26 | OSBPL9 | 2570373 | 2541 | 0.195 | 0.0639 | No | ||
| 27 | RAB3D | 4210253 | 2636 | 0.179 | 0.0613 | No | ||
| 28 | ENTPD4 | 7040288 | 2641 | 0.178 | 0.0635 | No | ||
| 29 | MIB1 | 3800537 | 2654 | 0.176 | 0.0652 | No | ||
| 30 | UBE2G1 | 2230546 5420528 7040463 | 2683 | 0.172 | 0.0661 | No | ||
| 31 | PCTK1 | 4560079 | 2767 | 0.159 | 0.0637 | No | ||
| 32 | ZBTB7A | 3780092 | 2800 | 0.153 | 0.0641 | No | ||
| 33 | SEMA4F | 3850056 | 2833 | 0.150 | 0.0644 | No | ||
| 34 | ANPEP | 6510138 | 2965 | 0.132 | 0.0590 | No | ||
| 35 | COPS7B | 2970484 4280022 | 2977 | 0.131 | 0.0602 | No | ||
| 36 | SLC7A1 | 2450193 2710241 | 3028 | 0.125 | 0.0592 | No | ||
| 37 | PHF15 | 6550066 6860673 | 3065 | 0.122 | 0.0589 | No | ||
| 38 | SLC7A6 | 60687 | 3090 | 0.119 | 0.0592 | No | ||
| 39 | TXNRD1 | 6590446 | 3117 | 0.115 | 0.0594 | No | ||
| 40 | EAF1 | 5420039 6550619 | 3146 | 0.113 | 0.0594 | No | ||
| 41 | STC1 | 360161 | 3185 | 0.109 | 0.0588 | No | ||
| 42 | ETS1 | 5270278 6450717 6620465 | 3268 | 0.100 | 0.0557 | No | ||
| 43 | ADAM9 | 3360411 | 3326 | 0.096 | 0.0539 | No | ||
| 44 | LRP4 | 1740253 | 3344 | 0.094 | 0.0543 | No | ||
| 45 | DYNLT3 | 5570398 | 3542 | 0.079 | 0.0447 | No | ||
| 46 | FAM62A | 5080008 | 3549 | 0.079 | 0.0454 | No | ||
| 47 | MAP3K10 | 3360168 4850180 | 3613 | 0.075 | 0.0430 | No | ||
| 48 | ABR | 610079 1170609 3610195 5670050 | 3737 | 0.068 | 0.0372 | No | ||
| 49 | ORC2L | 1990470 6510019 | 3784 | 0.065 | 0.0356 | No | ||
| 50 | E2F3 | 50162 460180 | 3800 | 0.064 | 0.0357 | No | ||
| 51 | APBA2BP | 5700341 | 3880 | 0.059 | 0.0322 | No | ||
| 52 | CNNM1 | 2190433 | 3911 | 0.058 | 0.0313 | No | ||
| 53 | PPP2R5C | 3310121 6650672 | 3930 | 0.057 | 0.0311 | No | ||
| 54 | ABHD6 | 630717 | 3960 | 0.056 | 0.0303 | No | ||
| 55 | HOXC6 | 3850133 4920736 | 4060 | 0.051 | 0.0256 | No | ||
| 56 | PODXL | 130717 | 4089 | 0.050 | 0.0248 | No | ||
| 57 | TGOLN2 | 4920433 | 4147 | 0.048 | 0.0223 | No | ||
| 58 | SYN2 | 3990593 | 4191 | 0.046 | 0.0206 | No | ||
| 59 | ZNF76 | 3850020 | 4264 | 0.044 | 0.0173 | No | ||
| 60 | KCNH7 | 7510068 | 4448 | 0.038 | 0.0079 | No | ||
| 61 | BAK1 | 6450563 | 4475 | 0.037 | 0.0070 | No | ||
| 62 | SOX11 | 610279 | 4515 | 0.036 | 0.0053 | No | ||
| 63 | PCTP | 6980324 | 4578 | 0.035 | 0.0024 | No | ||
| 64 | SGPL1 | 4480059 | 4660 | 0.032 | -0.0015 | No | ||
| 65 | RTN2 | 4480538 | 4723 | 0.031 | -0.0045 | No | ||
| 66 | MMP11 | 1980619 | 4747 | 0.030 | -0.0053 | No | ||
| 67 | FCHSD1 | 5080050 6450025 | 4838 | 0.028 | -0.0098 | No | ||
| 68 | IBRDC2 | 5130041 | 4868 | 0.028 | -0.0110 | No | ||
| 69 | SLITRK6 | 7320739 | 5040 | 0.025 | -0.0200 | No | ||
| 70 | NT5DC1 | 2320463 | 5161 | 0.023 | -0.0262 | No | ||
| 71 | SEMA4C | 1980315 | 5225 | 0.022 | -0.0293 | No | ||
| 72 | SMURF1 | 520609 580647 | 5292 | 0.021 | -0.0326 | No | ||
| 73 | MFN1 | 5080546 6200292 | 5307 | 0.021 | -0.0331 | No | ||
| 74 | ADD2 | 5910463 | 5434 | 0.020 | -0.0397 | No | ||
| 75 | RAB8B | 510692 1980017 3130136 4570739 | 5447 | 0.020 | -0.0401 | No | ||
| 76 | SNAI1 | 6590253 | 5501 | 0.019 | -0.0427 | No | ||
| 77 | SSTR3 | 5420064 | 5552 | 0.019 | -0.0452 | No | ||
| 78 | STARD13 | 540722 | 5844 | 0.016 | -0.0608 | No | ||
| 79 | MAP2K7 | 2260086 | 5971 | 0.015 | -0.0674 | No | ||
| 80 | IER2 | 2030008 | 6178 | 0.013 | -0.0785 | No | ||
| 81 | GALNT14 | 3990278 2030546 | 6182 | 0.013 | -0.0784 | No | ||
| 82 | HIVEP2 | 6520538 | 6207 | 0.013 | -0.0796 | No | ||
| 83 | ACCN2 | 6450465 | 6264 | 0.013 | -0.0824 | No | ||
| 84 | RNF121 | 110079 1580279 | 6270 | 0.013 | -0.0825 | No | ||
| 85 | MAMDC2 | 430471 | 6277 | 0.013 | -0.0827 | No | ||
| 86 | CD34 | 6650270 | 6422 | 0.012 | -0.0904 | No | ||
| 87 | RBM7 | 5890348 | 6743 | 0.010 | -0.1076 | No | ||
| 88 | CGN | 5890139 6370167 6400537 | 6781 | 0.010 | -0.1095 | No | ||
| 89 | DLL4 | 6400403 | 6873 | 0.009 | -0.1143 | No | ||
| 90 | UBE2R2 | 1230504 | 6951 | 0.009 | -0.1184 | No | ||
| 91 | SLC4A10 | 2060670 | 7003 | 0.009 | -0.1210 | No | ||
| 92 | OLFML2A | 5890039 | 7522 | 0.007 | -0.1491 | No | ||
| 93 | PLXNA1 | 70066 | 7596 | 0.007 | -0.1530 | No | ||
| 94 | MAP6 | 6860195 | 7602 | 0.007 | -0.1532 | No | ||
| 95 | TBX4 | 3060008 | 7671 | 0.006 | -0.1568 | No | ||
| 96 | SLC17A7 | 7000037 | 7672 | 0.006 | -0.1567 | No | ||
| 97 | BSN | 5270347 | 7723 | 0.006 | -0.1594 | No | ||
| 98 | PHOX2B | 5270075 | 7890 | 0.006 | -0.1683 | No | ||
| 99 | SLC4A4 | 1780075 1850040 4810068 | 8010 | 0.005 | -0.1747 | No | ||
| 100 | M6PR | 2480458 3290593 3840324 | 8237 | 0.004 | -0.1870 | No | ||
| 101 | GRIN2A | 6550538 | 8598 | 0.003 | -0.2065 | No | ||
| 102 | DVL3 | 360156 5390075 | 8751 | 0.003 | -0.2147 | No | ||
| 103 | DPF2 | 6980373 | 9631 | 0.001 | -0.2625 | No | ||
| 104 | GALNT7 | 3990280 | 9669 | 0.000 | -0.2646 | No | ||
| 105 | NIN | 3450576 5080048 | 9686 | 0.000 | -0.2654 | No | ||
| 106 | GPC4 | 5910408 | 9860 | -0.000 | -0.2748 | No | ||
| 107 | HOXB3 | 520292 | 9888 | -0.000 | -0.2763 | No | ||
| 108 | HMGB3 | 2940168 | 9953 | -0.000 | -0.2798 | No | ||
| 109 | MYLK | 4010600 7000364 | 10133 | -0.001 | -0.2895 | No | ||
| 110 | TOR2A | 1400026 | 10185 | -0.001 | -0.2923 | No | ||
| 111 | ABHD3 | 2370091 | 10318 | -0.002 | -0.2994 | No | ||
| 112 | SET | 6650286 | 10417 | -0.002 | -0.3047 | No | ||
| 113 | MYT1 | 6770524 | 10449 | -0.002 | -0.3064 | No | ||
| 114 | DIRAS1 | 2640270 | 10495 | -0.002 | -0.3088 | No | ||
| 115 | ATOH8 | 4540204 | 10526 | -0.002 | -0.3104 | No | ||
| 116 | SATB2 | 2470181 | 10578 | -0.002 | -0.3132 | No | ||
| 117 | KCNS3 | 4150039 | 10644 | -0.002 | -0.3167 | No | ||
| 118 | GRB10 | 6980082 | 10821 | -0.003 | -0.3262 | No | ||
| 119 | RHOQ | 520161 | 11000 | -0.003 | -0.3358 | No | ||
| 120 | EIF5A2 | 7040139 | 11261 | -0.004 | -0.3499 | No | ||
| 121 | VDR | 130156 510438 | 11278 | -0.004 | -0.3507 | No | ||
| 122 | LIFR | 3360068 3830102 | 11313 | -0.005 | -0.3525 | No | ||
| 123 | PHYHIP | 1170377 | 11383 | -0.005 | -0.3562 | No | ||
| 124 | SH3BP4 | 3140242 | 11409 | -0.005 | -0.3575 | No | ||
| 125 | VPS4B | 3440332 | 11477 | -0.005 | -0.3611 | No | ||
| 126 | LYPLA2 | 1500112 | 11689 | -0.006 | -0.3725 | No | ||
| 127 | SLC8A2 | 6220092 | 11737 | -0.006 | -0.3750 | No | ||
| 128 | CCNJ | 870047 | 11748 | -0.006 | -0.3754 | No | ||
| 129 | ATP1B4 | 5670546 | 11779 | -0.006 | -0.3770 | No | ||
| 130 | NCOR2 | 1980575 6550050 | 11835 | -0.006 | -0.3799 | No | ||
| 131 | PRDM1 | 3170347 3520301 | 11997 | -0.007 | -0.3885 | No | ||
| 132 | RET | 3850746 | 12008 | -0.007 | -0.3890 | No | ||
| 133 | DOCK3 | 3290273 3360497 2850068 | 12070 | -0.007 | -0.3922 | No | ||
| 134 | SLC39A9 | 3940348 | 12136 | -0.008 | -0.3956 | No | ||
| 135 | DDX42 | 2060546 | 12304 | -0.008 | -0.4046 | No | ||
| 136 | PAPOLA | 4200286 4850112 | 12858 | -0.011 | -0.4345 | No | ||
| 137 | CPEB3 | 3940164 | 12861 | -0.011 | -0.4345 | No | ||
| 138 | BAI1 | 6350053 | 13043 | -0.013 | -0.4442 | No | ||
| 139 | BMPR2 | 5340066 6960328 | 13057 | -0.013 | -0.4447 | No | ||
| 140 | LRFN2 | 6450411 | 13164 | -0.014 | -0.4503 | No | ||
| 141 | HCN4 | 1050026 | 13172 | -0.014 | -0.4505 | No | ||
| 142 | GOPC | 6620400 | 13181 | -0.014 | -0.4507 | No | ||
| 143 | GGTL3 | 1770288 | 13198 | -0.014 | -0.4514 | No | ||
| 144 | ABCC5 | 2100600 5050692 | 13549 | -0.017 | -0.4702 | No | ||
| 145 | ASB13 | 1090070 | 13698 | -0.019 | -0.4780 | No | ||
| 146 | USP37 | 2470433 | 13701 | -0.019 | -0.4779 | No | ||
| 147 | MASP1 | 1780619 2900066 | 13748 | -0.019 | -0.4801 | No | ||
| 148 | PAFAH1B1 | 4230333 6420121 6450066 | 14002 | -0.023 | -0.4936 | No | ||
| 149 | LNPEP | 1170092 1940093 3120025 | 14085 | -0.024 | -0.4977 | No | ||
| 150 | NIPA1 | 380600 | 14166 | -0.026 | -0.5017 | No | ||
| 151 | MKNK2 | 1240020 5270092 | 14508 | -0.032 | -0.5198 | No | ||
| 152 | CSNK2A1 | 1580577 | 14579 | -0.034 | -0.5232 | No | ||
| 153 | MTUS1 | 780348 4920609 | 14792 | -0.042 | -0.5341 | No | ||
| 154 | GGA2 | 520451 1240082 2650332 | 14803 | -0.042 | -0.5341 | No | ||
| 155 | FKHL18 | 6370603 | 14912 | -0.047 | -0.5393 | No | ||
| 156 | ST8SIA4 | 2680605 3060215 | 15082 | -0.057 | -0.5477 | No | ||
| 157 | CACNB1 | 2940427 3710487 | 15112 | -0.059 | -0.5485 | No | ||
| 158 | ADAM11 | 1050008 3130494 | 15197 | -0.065 | -0.5522 | No | ||
| 159 | ITGB3 | 5270463 | 15243 | -0.068 | -0.5537 | No | ||
| 160 | ASB6 | 2570600 3850092 | 15285 | -0.071 | -0.5550 | No | ||
| 161 | SLC27A4 | 4060139 | 15333 | -0.074 | -0.5565 | No | ||
| 162 | FBXW4 | 3060577 5670095 | 15347 | -0.075 | -0.5562 | No | ||
| 163 | ZNF385 | 450176 4810358 | 15405 | -0.080 | -0.5582 | No | ||
| 164 | USP12 | 6130735 6620280 | 15437 | -0.083 | -0.5588 | No | ||
| 165 | SLC35A4 | 2940671 | 15473 | -0.086 | -0.5595 | No | ||
| 166 | SUV39H1 | 630112 | 15532 | -0.092 | -0.5614 | No | ||
| 167 | KLC2 | 1240048 5340441 | 15576 | -0.098 | -0.5624 | No | ||
| 168 | CORO2A | 3840605 6620390 | 15682 | -0.109 | -0.5666 | No | ||
| 169 | NRM | 520068 | 15693 | -0.109 | -0.5657 | No | ||
| 170 | SAMD10 | 1580497 | 15784 | -0.119 | -0.5690 | No | ||
| 171 | TBC1D1 | 2510072 | 15851 | -0.126 | -0.5708 | No | ||
| 172 | DICER1 | 540286 | 15855 | -0.127 | -0.5693 | No | ||
| 173 | ZFYVE1 | 60440 | 15937 | -0.135 | -0.5718 | No | ||
| 174 | ETV6 | 610524 | 16017 | -0.145 | -0.5742 | No | ||
| 175 | SORT1 | 1850242 2320632 3870537 | 16037 | -0.148 | -0.5732 | No | ||
| 176 | UBN1 | 5900195 | 16075 | -0.155 | -0.5731 | No | ||
| 177 | RHOT2 | 1660053 2120292 | 16158 | -0.166 | -0.5753 | No | ||
| 178 | ATXN1 | 5550156 | 16338 | -0.192 | -0.5824 | No | ||
| 179 | PELI2 | 1940364 | 16432 | -0.210 | -0.5846 | Yes | ||
| 180 | PARP14 | 5860487 | 16449 | -0.213 | -0.5826 | Yes | ||
| 181 | MAPK14 | 5290731 | 16518 | -0.228 | -0.5831 | Yes | ||
| 182 | LFNG | 5360711 | 16528 | -0.231 | -0.5805 | Yes | ||
| 183 | UBTD1 | 6900315 | 16537 | -0.234 | -0.5777 | Yes | ||
| 184 | ESRRA | 6550242 | 16573 | -0.242 | -0.5763 | Yes | ||
| 185 | SMARCD2 | 3830619 | 16579 | -0.244 | -0.5733 | Yes | ||
| 186 | PTPN18 | 2640537 | 16630 | -0.254 | -0.5725 | Yes | ||
| 187 | SUV420H2 | 4050368 | 16766 | -0.285 | -0.5760 | Yes | ||
| 188 | IRF4 | 610133 | 16775 | -0.287 | -0.5725 | Yes | ||
| 189 | OAZ2 | 2810110 | 16802 | -0.294 | -0.5699 | Yes | ||
| 190 | TRAPPC6B | 5080184 5290068 | 16964 | -0.338 | -0.5741 | Yes | ||
| 191 | RFXANK | 4010341 | 17040 | -0.359 | -0.5732 | Yes | ||
| 192 | BRPF1 | 3170139 | 17066 | -0.370 | -0.5696 | Yes | ||
| 193 | SIRT7 | 5890632 | 17102 | -0.385 | -0.5662 | Yes | ||
| 194 | HIC2 | 4010433 | 17104 | -0.385 | -0.5610 | Yes | ||
| 195 | ATP10D | 4850139 | 17119 | -0.391 | -0.5564 | Yes | ||
| 196 | PHC2 | 1430433 | 17164 | -0.403 | -0.5533 | Yes | ||
| 197 | BCL9L | 6940053 | 17235 | -0.427 | -0.5513 | Yes | ||
| 198 | CASP2 | 2260692 | 17242 | -0.430 | -0.5458 | Yes | ||
| 199 | TLK2 | 1170161 | 17304 | -0.455 | -0.5429 | Yes | ||
| 200 | SEMA4D | 7050152 | 17314 | -0.460 | -0.5371 | Yes | ||
| 201 | SEMA4B | 110347 | 17403 | -0.498 | -0.5351 | Yes | ||
| 202 | CASC3 | 5890095 | 17418 | -0.502 | -0.5290 | Yes | ||
| 203 | PCQAP | 3060064 6200273 | 17476 | -0.530 | -0.5249 | Yes | ||
| 204 | PCSK7 | 6370450 | 17497 | -0.538 | -0.5186 | Yes | ||
| 205 | MTMR3 | 1050014 4920180 | 17564 | -0.570 | -0.5144 | Yes | ||
| 206 | BAZ2A | 730184 | 17621 | -0.608 | -0.5092 | Yes | ||
| 207 | DNAJB5 | 2900215 | 17628 | -0.611 | -0.5012 | Yes | ||
| 208 | DCTN1 | 1450056 6590022 | 17630 | -0.611 | -0.4929 | Yes | ||
| 209 | MXD4 | 4810113 | 17661 | -0.632 | -0.4859 | Yes | ||
| 210 | MYO18A | 1230333 | 17664 | -0.634 | -0.4773 | Yes | ||
| 211 | PPP1CA | 1450722 3140181 1770133 4070092 | 17702 | -0.656 | -0.4704 | Yes | ||
| 212 | ANKRD50 | 870543 | 17719 | -0.668 | -0.4622 | Yes | ||
| 213 | WARS | 2570717 3290139 | 17753 | -0.685 | -0.4546 | Yes | ||
| 214 | SYVN1 | 6940594 | 17809 | -0.720 | -0.4478 | Yes | ||
| 215 | MAP3K11 | 7000039 | 17815 | -0.725 | -0.4382 | Yes | ||
| 216 | ABTB1 | 4780086 | 17858 | -0.754 | -0.4302 | Yes | ||
| 217 | SERTAD3 | 2810088 | 17862 | -0.754 | -0.4200 | Yes | ||
| 218 | STAT3 | 460040 3710341 | 18021 | -0.893 | -0.4164 | Yes | ||
| 219 | RNF40 | 3610397 | 18034 | -0.906 | -0.4047 | Yes | ||
| 220 | CDC42SE1 | 510037 3780438 | 18087 | -0.974 | -0.3943 | Yes | ||
| 221 | FLOT2 | 50358 4210068 5080397 | 18102 | -0.989 | -0.3816 | Yes | ||
| 222 | LBH | 70093 | 18110 | -0.999 | -0.3683 | Yes | ||
| 223 | USP52 | 3710215 6380133 | 18151 | -1.047 | -0.3562 | Yes | ||
| 224 | RNF44 | 5910692 | 18193 | -1.116 | -0.3432 | Yes | ||
| 225 | PTPRF | 1770528 3190044 | 18211 | -1.146 | -0.3285 | Yes | ||
| 226 | IHPK1 | 1340053 | 18237 | -1.184 | -0.3137 | Yes | ||
| 227 | SCARB1 | 510441 6770047 | 18239 | -1.188 | -0.2975 | Yes | ||
| 228 | KLF13 | 4670563 | 18251 | -1.209 | -0.2816 | Yes | ||
| 229 | CBX7 | 3940035 | 18268 | -1.266 | -0.2652 | Yes | ||
| 230 | ZSWIM4 | 6940195 | 18308 | -1.344 | -0.2490 | Yes | ||
| 231 | GTPBP2 | 2760750 | 18310 | -1.346 | -0.2307 | Yes | ||
| 232 | MCL1 | 1660672 | 18315 | -1.362 | -0.2123 | Yes | ||
| 233 | PLAGL2 | 1770148 | 18356 | -1.489 | -0.1942 | Yes | ||
| 234 | ARID3B | 6650008 | 18476 | -1.964 | -0.1739 | Yes | ||
| 235 | ST6GAL1 | 2970546 | 18481 | -2.005 | -0.1467 | Yes | ||
| 236 | TEF | 1050050 3120397 4060487 | 18538 | -2.509 | -0.1156 | Yes | ||
| 237 | EIF4EBP1 | 60132 5720148 | 18539 | -2.520 | -0.0812 | Yes | ||
| 238 | IL16 | 3850168 | 18571 | -2.923 | -0.0430 | Yes | ||
| 239 | SRF | 70139 2320039 | 18588 | -3.326 | 0.0015 | Yes |

