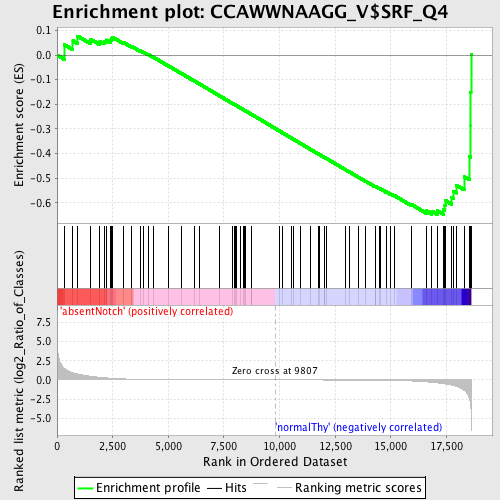
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CCAWWNAAGG_V$SRF_Q4 |
| Enrichment Score (ES) | -0.64703494 |
| Normalized Enrichment Score (NES) | -1.4650818 |
| Nominal p-value | 0.019753087 |
| FDR q-value | 0.26483908 |
| FWER p-Value | 0.922 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ACTB | 1660736 2030204 2060215 5050047 | 326 | 1.476 | 0.0415 | No | ||
| 2 | MBNL1 | 2640762 7100048 | 713 | 0.914 | 0.0573 | No | ||
| 3 | CKM | 1450524 | 924 | 0.765 | 0.0766 | No | ||
| 4 | KCTD15 | 2810735 | 1516 | 0.469 | 0.0635 | No | ||
| 5 | CORO1C | 4570397 | 1922 | 0.339 | 0.0553 | No | ||
| 6 | MAPKAPK2 | 2850500 | 2120 | 0.284 | 0.0561 | No | ||
| 7 | PLCB3 | 4670402 | 2223 | 0.258 | 0.0609 | No | ||
| 8 | TCF4 | 520021 | 2397 | 0.223 | 0.0605 | No | ||
| 9 | DAPK3 | 4590121 | 2455 | 0.213 | 0.0660 | No | ||
| 10 | H3F3A | 2900086 | 2485 | 0.208 | 0.0727 | No | ||
| 11 | TPM2 | 520735 3870390 | 2996 | 0.128 | 0.0504 | No | ||
| 12 | FGFRL1 | 6370047 | 3337 | 0.095 | 0.0358 | No | ||
| 13 | SCOC | 610048 2230053 | 3742 | 0.067 | 0.0168 | No | ||
| 14 | NPAS2 | 770670 | 3895 | 0.059 | 0.0109 | No | ||
| 15 | MRVI1 | 4810338 4850601 5900441 | 4090 | 0.050 | 0.0025 | No | ||
| 16 | PCDH7 | 450398 4150040 | 4314 | 0.042 | -0.0079 | No | ||
| 17 | MYL7 | 2480541 | 4984 | 0.026 | -0.0429 | No | ||
| 18 | PICALM | 1940280 | 5596 | 0.018 | -0.0751 | No | ||
| 19 | TNNC1 | 1990575 | 6161 | 0.014 | -0.1050 | No | ||
| 20 | MYH11 | 7100273 | 6381 | 0.012 | -0.1163 | No | ||
| 21 | NKX2-2 | 4150731 | 7316 | 0.008 | -0.1663 | No | ||
| 22 | PODN | 3710068 | 7878 | 0.006 | -0.1964 | No | ||
| 23 | ATRX | 2340039 3610132 4210373 | 7963 | 0.005 | -0.2007 | No | ||
| 24 | ERBB2IP | 580253 1090672 | 8001 | 0.005 | -0.2025 | No | ||
| 25 | TITF1 | 3390554 4540292 | 8062 | 0.005 | -0.2055 | No | ||
| 26 | KCNK3 | 2690372 | 8259 | 0.004 | -0.2159 | No | ||
| 27 | CALD1 | 1770129 1940397 | 8364 | 0.004 | -0.2213 | No | ||
| 28 | HOXD13 | 2260603 | 8438 | 0.004 | -0.2251 | No | ||
| 29 | HOXC5 | 1580017 | 8463 | 0.004 | -0.2262 | No | ||
| 30 | SDPR | 3360292 | 8723 | 0.003 | -0.2401 | No | ||
| 31 | HOXD10 | 6100039 | 8737 | 0.003 | -0.2407 | No | ||
| 32 | THBS1 | 4560494 430288 | 10006 | -0.001 | -0.3090 | No | ||
| 33 | MYLK | 4010600 7000364 | 10133 | -0.001 | -0.3157 | No | ||
| 34 | NPPA | 6550328 | 10542 | -0.002 | -0.3377 | No | ||
| 35 | NR4A1 | 6290161 | 10638 | -0.002 | -0.3427 | No | ||
| 36 | TAGLN | 6660280 | 10929 | -0.003 | -0.3582 | No | ||
| 37 | NKX6-1 | 6040731 | 11368 | -0.005 | -0.3816 | No | ||
| 38 | DIXDC1 | 6980435 | 11743 | -0.006 | -0.4015 | No | ||
| 39 | EPHB1 | 4610056 | 11784 | -0.006 | -0.4034 | No | ||
| 40 | PRDM1 | 3170347 3520301 | 11997 | -0.007 | -0.4146 | No | ||
| 41 | SP7 | 1050315 | 12002 | -0.007 | -0.4145 | No | ||
| 42 | VIT | 6620301 | 12096 | -0.008 | -0.4192 | No | ||
| 43 | FOSB | 1940142 | 12947 | -0.012 | -0.4646 | No | ||
| 44 | MRGPRF | 4780746 | 13151 | -0.013 | -0.4750 | No | ||
| 45 | RARB | 430139 1410138 | 13554 | -0.017 | -0.4960 | No | ||
| 46 | TRDN | 1580411 | 13879 | -0.021 | -0.5126 | No | ||
| 47 | SVIL | 2450347 4050735 4070576 4120706 5390161 5860142 6520278 7100397 | 14300 | -0.028 | -0.5341 | No | ||
| 48 | ACTA1 | 840538 | 14312 | -0.028 | -0.5336 | No | ||
| 49 | SCN3B | 840112 1780152 5910685 6100131 | 14486 | -0.032 | -0.5416 | No | ||
| 50 | PLN | 3940132 | 14523 | -0.033 | -0.5423 | No | ||
| 51 | CFL2 | 380239 6200368 | 14783 | -0.041 | -0.5546 | No | ||
| 52 | PPP1R12A | 4070066 4590162 | 14994 | -0.052 | -0.5638 | No | ||
| 53 | INSM1 | 3450671 | 15151 | -0.061 | -0.5698 | No | ||
| 54 | PFN1 | 6130132 | 15921 | -0.133 | -0.6059 | No | ||
| 55 | UBE2C | 6130017 | 16589 | -0.245 | -0.6320 | No | ||
| 56 | ANP32E | 6510706 | 16847 | -0.306 | -0.6337 | Yes | ||
| 57 | NPM3 | 3610300 | 17096 | -0.382 | -0.6317 | Yes | ||
| 58 | ACTG2 | 4780180 | 17378 | -0.486 | -0.6274 | Yes | ||
| 59 | RUNDC1 | 2450037 | 17430 | -0.508 | -0.6098 | Yes | ||
| 60 | PHF12 | 870400 3130687 | 17447 | -0.516 | -0.5900 | Yes | ||
| 61 | FLNA | 5390193 | 17743 | -0.680 | -0.5787 | Yes | ||
| 62 | DUSP2 | 2370184 | 17814 | -0.722 | -0.5535 | Yes | ||
| 63 | CSRP1 | 2810403 | 17934 | -0.811 | -0.5275 | Yes | ||
| 64 | EGR2 | 3800403 | 18301 | -1.337 | -0.4936 | Yes | ||
| 65 | RASSF2 | 2480079 5720546 | 18520 | -2.347 | -0.4114 | Yes | ||
| 66 | LSP1 | 2350091 | 18586 | -3.268 | -0.2840 | Yes | ||
| 67 | SRF | 70139 2320039 | 18588 | -3.326 | -0.1508 | Yes | ||
| 68 | EGR1 | 4610347 | 18604 | -3.801 | 0.0006 | Yes |

