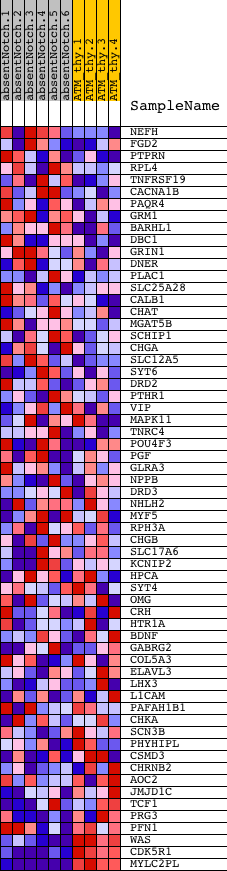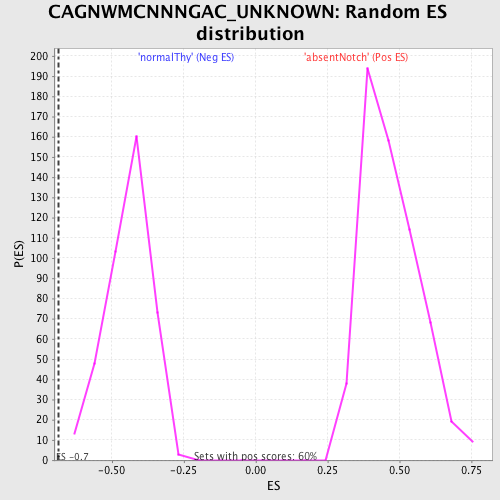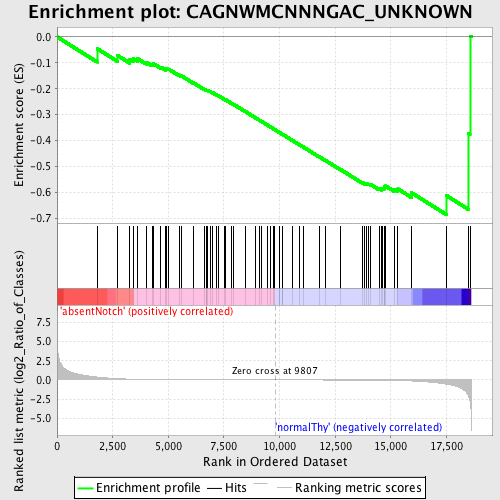
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CAGNWMCNNNGAC_UNKNOWN |
| Enrichment Score (ES) | -0.6846136 |
| Normalized Enrichment Score (NES) | -1.54918 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.19833249 |
| FWER p-Value | 0.552 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NEFH | 630239 | 1798 | 0.376 | -0.0452 | No | ||
| 2 | FGD2 | 5340195 | 2707 | 0.167 | -0.0711 | No | ||
| 3 | PTPRN | 5900577 | 3270 | 0.100 | -0.0876 | No | ||
| 4 | RPL4 | 2940014 5420324 | 3429 | 0.087 | -0.0841 | No | ||
| 5 | TNFRSF19 | 3870731 6940730 | 3626 | 0.074 | -0.0845 | No | ||
| 6 | CACNA1B | 1740010 5360593 | 4002 | 0.054 | -0.0973 | No | ||
| 7 | PAQR4 | 730736 | 4275 | 0.043 | -0.1061 | No | ||
| 8 | GRM1 | 5890324 | 4329 | 0.042 | -0.1032 | No | ||
| 9 | BARHL1 | 4210113 | 4661 | 0.032 | -0.1165 | No | ||
| 10 | DBC1 | 4610156 | 4888 | 0.028 | -0.1249 | No | ||
| 11 | GRIN1 | 3800014 7000609 | 4930 | 0.027 | -0.1234 | No | ||
| 12 | DNER | 3610170 | 5002 | 0.026 | -0.1237 | No | ||
| 13 | PLAC1 | 2350594 | 5480 | 0.019 | -0.1468 | No | ||
| 14 | SLC25A28 | 6590600 | 5579 | 0.018 | -0.1496 | No | ||
| 15 | CALB1 | 460070 | 6109 | 0.014 | -0.1762 | No | ||
| 16 | CHAT | 6840603 | 6638 | 0.011 | -0.2031 | No | ||
| 17 | MGAT5B | 380131 2570364 | 6698 | 0.010 | -0.2049 | No | ||
| 18 | SCHIP1 | 4560152 | 6722 | 0.010 | -0.2047 | No | ||
| 19 | CHGA | 1990056 6550463 | 6753 | 0.010 | -0.2049 | No | ||
| 20 | SLC12A5 | 1980692 | 6881 | 0.009 | -0.2105 | No | ||
| 21 | SYT6 | 2510280 3850128 4540064 | 6997 | 0.009 | -0.2154 | No | ||
| 22 | DRD2 | 5890369 | 7145 | 0.008 | -0.2222 | No | ||
| 23 | PTHR1 | 2640711 3360647 | 7234 | 0.008 | -0.2259 | No | ||
| 24 | VIP | 2850647 | 7543 | 0.007 | -0.2415 | No | ||
| 25 | MAPK11 | 130452 3120440 3610465 | 7589 | 0.007 | -0.2430 | No | ||
| 26 | TNRC4 | 4050156 | 7834 | 0.006 | -0.2554 | No | ||
| 27 | POU4F3 | 2690035 | 7925 | 0.005 | -0.2595 | No | ||
| 28 | PGF | 4810593 | 8478 | 0.004 | -0.2887 | No | ||
| 29 | GLRA3 | 2570500 | 8896 | 0.003 | -0.3108 | No | ||
| 30 | NPPB | 4150722 | 9104 | 0.002 | -0.3217 | No | ||
| 31 | DRD3 | 4780402 | 9191 | 0.002 | -0.3261 | No | ||
| 32 | NHLH2 | 6200021 | 9445 | 0.001 | -0.3396 | No | ||
| 33 | MYF5 | 1050739 | 9573 | 0.001 | -0.3463 | No | ||
| 34 | RPH3A | 2190156 | 9738 | 0.000 | -0.3552 | No | ||
| 35 | CHGB | 1980053 | 9767 | 0.000 | -0.3566 | No | ||
| 36 | SLC17A6 | 4210576 | 10000 | -0.001 | -0.3691 | No | ||
| 37 | KCNIP2 | 60088 1780324 | 10004 | -0.001 | -0.3691 | No | ||
| 38 | HPCA | 6860010 | 10122 | -0.001 | -0.3753 | No | ||
| 39 | SYT4 | 5080193 | 10149 | -0.001 | -0.3766 | No | ||
| 40 | OMG | 6760066 | 10592 | -0.002 | -0.4001 | No | ||
| 41 | CRH | 3710301 | 10901 | -0.003 | -0.4162 | No | ||
| 42 | HTR1A | 6980441 | 11053 | -0.004 | -0.4239 | No | ||
| 43 | BDNF | 2940128 3520368 | 11795 | -0.006 | -0.4630 | No | ||
| 44 | GABRG2 | 2350402 4210204 6130279 6550037 | 12053 | -0.007 | -0.4758 | No | ||
| 45 | COL5A3 | 1940471 | 12747 | -0.011 | -0.5117 | No | ||
| 46 | ELAVL3 | 2850014 | 13742 | -0.019 | -0.5626 | No | ||
| 47 | LHX3 | 670164 1770280 | 13835 | -0.020 | -0.5647 | No | ||
| 48 | L1CAM | 4850021 | 13895 | -0.021 | -0.5650 | No | ||
| 49 | PAFAH1B1 | 4230333 6420121 6450066 | 14002 | -0.023 | -0.5675 | No | ||
| 50 | CHKA | 510324 | 14104 | -0.024 | -0.5696 | No | ||
| 51 | SCN3B | 840112 1780152 5910685 6100131 | 14486 | -0.032 | -0.5858 | No | ||
| 52 | PHYHIPL | 2360706 3840692 | 14575 | -0.034 | -0.5858 | No | ||
| 53 | CSMD3 | 1660427 1940687 | 14608 | -0.035 | -0.5826 | No | ||
| 54 | CHRNB2 | 580204 3120739 | 14695 | -0.038 | -0.5821 | No | ||
| 55 | AOC2 | 520348 3120008 | 14723 | -0.039 | -0.5781 | No | ||
| 56 | JMJD1C | 940575 2120025 | 14762 | -0.040 | -0.5746 | No | ||
| 57 | TCF1 | 5390022 | 15147 | -0.061 | -0.5869 | No | ||
| 58 | PRG3 | 6220020 | 15321 | -0.073 | -0.5862 | No | ||
| 59 | PFN1 | 6130132 | 15921 | -0.133 | -0.6003 | No | ||
| 60 | WAS | 5270193 | 17487 | -0.535 | -0.6110 | Yes | ||
| 61 | CDK5R1 | 3870161 | 18498 | -2.123 | -0.3733 | Yes | ||
| 62 | MYLC2PL | 360280 | 18560 | -2.758 | 0.0030 | Yes |