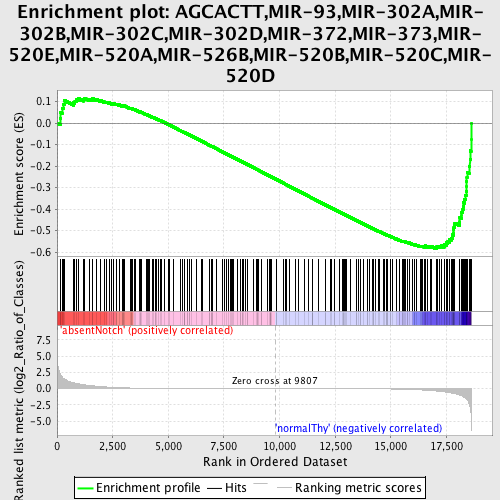
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | AGCACTT,MIR-93,MIR-302A,MIR-302B,MIR-302C,MIR-302D,MIR-372,MIR-373,MIR-520E,MIR-520A,MIR-526B,MIR-520B,MIR-520C,MIR-520D |
| Enrichment Score (ES) | -0.5857175 |
| Normalized Enrichment Score (NES) | -1.5499711 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.23544829 |
| FWER p-Value | 0.55 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP3R1 | 1190041 1570487 | 140 | 2.264 | 0.0225 | No | ||
| 2 | RGMA | 2450711 | 164 | 2.113 | 0.0493 | No | ||
| 3 | INHBB | 2680092 | 227 | 1.782 | 0.0697 | No | ||
| 4 | SLC6A9 | 2470520 | 276 | 1.588 | 0.0882 | No | ||
| 5 | LEFTY1 | 3290452 | 319 | 1.486 | 0.1056 | No | ||
| 6 | APP | 2510053 | 730 | 0.900 | 0.0953 | No | ||
| 7 | RBBP7 | 430113 450450 2370309 | 801 | 0.838 | 0.1026 | No | ||
| 8 | MRPS25 | 610441 870128 1410368 | 881 | 0.788 | 0.1088 | No | ||
| 9 | SDC1 | 3440471 6350408 | 962 | 0.741 | 0.1143 | No | ||
| 10 | TNFAIP1 | 6940593 | 1181 | 0.614 | 0.1106 | No | ||
| 11 | NEK9 | 3440402 | 1243 | 0.584 | 0.1151 | No | ||
| 12 | CCND1 | 460524 770309 3120576 6980398 | 1439 | 0.499 | 0.1111 | No | ||
| 13 | RPS6KA1 | 4120647 | 1583 | 0.443 | 0.1092 | No | ||
| 14 | CUGBP2 | 6180121 6840139 | 1584 | 0.443 | 0.1151 | No | ||
| 15 | CXXC6 | 2810168 | 1791 | 0.380 | 0.1090 | No | ||
| 16 | AK2 | 4590458 4760195 6110097 | 1935 | 0.334 | 0.1056 | No | ||
| 17 | DAZAP2 | 6200102 | 2141 | 0.281 | 0.0982 | No | ||
| 18 | RPS6KA3 | 1980707 | 2228 | 0.258 | 0.0970 | No | ||
| 19 | SENP1 | 6100537 | 2340 | 0.234 | 0.0941 | No | ||
| 20 | TRPS1 | 870100 | 2453 | 0.213 | 0.0908 | No | ||
| 21 | YWHAZ | 1230717 | 2520 | 0.199 | 0.0899 | No | ||
| 22 | CDCA7 | 3060097 | 2549 | 0.194 | 0.0909 | No | ||
| 23 | RAB22A | 110500 3830707 | 2671 | 0.174 | 0.0867 | No | ||
| 24 | TRIM36 | 1400600 1980082 5130037 | 2685 | 0.172 | 0.0882 | No | ||
| 25 | RBL1 | 3130372 | 2784 | 0.156 | 0.0850 | No | ||
| 26 | YTHDF3 | 4540632 4670717 7040609 | 2938 | 0.134 | 0.0785 | No | ||
| 27 | MARCH8 | 60397 | 2951 | 0.133 | 0.0796 | No | ||
| 28 | NEO1 | 1990154 | 2967 | 0.132 | 0.0805 | No | ||
| 29 | ASF1A | 7050020 | 2973 | 0.131 | 0.0820 | No | ||
| 30 | PBX3 | 1300424 3710577 6180575 | 3026 | 0.126 | 0.0808 | No | ||
| 31 | NR4A2 | 60273 | 3289 | 0.098 | 0.0679 | No | ||
| 32 | ADAM9 | 3360411 | 3326 | 0.096 | 0.0672 | No | ||
| 33 | ZFP36L2 | 510180 | 3354 | 0.094 | 0.0670 | No | ||
| 34 | SS18L1 | 4610180 6550068 | 3404 | 0.090 | 0.0655 | No | ||
| 35 | PFN2 | 730040 5700026 | 3473 | 0.084 | 0.0629 | No | ||
| 36 | RDBP | 6590044 | 3535 | 0.079 | 0.0607 | No | ||
| 37 | SAR1B | 2510273 | 3710 | 0.069 | 0.0521 | No | ||
| 38 | DCUN1D1 | 2360332 | 3725 | 0.068 | 0.0523 | No | ||
| 39 | KPNA2 | 3450114 | 3762 | 0.066 | 0.0512 | No | ||
| 40 | ARID4B | 730754 6660026 | 3777 | 0.065 | 0.0513 | No | ||
| 41 | RABGAP1 | 5050397 | 4021 | 0.053 | 0.0388 | No | ||
| 42 | MBNL2 | 3450707 | 4040 | 0.052 | 0.0385 | No | ||
| 43 | ANKRD13C | 5270202 | 4100 | 0.050 | 0.0359 | No | ||
| 44 | RHOC | 2060242 | 4130 | 0.048 | 0.0350 | No | ||
| 45 | RNF6 | 2640035 | 4298 | 0.043 | 0.0265 | No | ||
| 46 | POLQ | 1050053 2190102 | 4341 | 0.041 | 0.0248 | No | ||
| 47 | EDNRB | 4280717 6400435 | 4421 | 0.038 | 0.0210 | No | ||
| 48 | BRMS1L | 6110609 | 4438 | 0.038 | 0.0206 | No | ||
| 49 | PCDHA11 | 1510253 | 4483 | 0.037 | 0.0187 | No | ||
| 50 | ST8SIA2 | 2680082 | 4545 | 0.035 | 0.0159 | No | ||
| 51 | ESR1 | 4060372 5860193 | 4638 | 0.033 | 0.0113 | No | ||
| 52 | NFIB | 460450 | 4673 | 0.032 | 0.0099 | No | ||
| 53 | DPYSL5 | 4900333 | 4814 | 0.029 | 0.0027 | No | ||
| 54 | ETV1 | 70735 2940603 5080463 | 5007 | 0.026 | -0.0074 | No | ||
| 55 | LASS6 | 5890576 6520138 | 5034 | 0.025 | -0.0085 | No | ||
| 56 | PCDHA6 | 780333 2370280 | 5229 | 0.022 | -0.0188 | No | ||
| 57 | DRD1 | 430025 | 5550 | 0.019 | -0.0359 | No | ||
| 58 | PURB | 5360138 | 5614 | 0.018 | -0.0391 | No | ||
| 59 | RABEP1 | 4760427 6200463 | 5712 | 0.017 | -0.0442 | No | ||
| 60 | ARHGEF10 | 3120593 6900673 | 5717 | 0.017 | -0.0442 | No | ||
| 61 | LATS2 | 4480593 6020494 | 5726 | 0.017 | -0.0444 | No | ||
| 62 | RALGDS | 430463 | 5840 | 0.016 | -0.0503 | No | ||
| 63 | UBE2J1 | 4070133 | 5881 | 0.016 | -0.0523 | No | ||
| 64 | FNDC3A | 2690097 3290044 | 5935 | 0.015 | -0.0549 | No | ||
| 65 | ZFPM2 | 460072 | 5951 | 0.015 | -0.0556 | No | ||
| 66 | OLFM3 | 580112 1050072 4070204 | 6022 | 0.015 | -0.0592 | No | ||
| 67 | MLLT6 | 3870168 | 6281 | 0.013 | -0.0730 | No | ||
| 68 | SYNC1 | 580114 | 6507 | 0.011 | -0.0851 | No | ||
| 69 | FLT1 | 3830167 4920438 | 6514 | 0.011 | -0.0853 | No | ||
| 70 | PHF6 | 3120632 3990041 | 6832 | 0.010 | -0.1024 | No | ||
| 71 | NPAS3 | 630408 4610066 | 6836 | 0.010 | -0.1024 | No | ||
| 72 | TIPARP | 50397 | 6921 | 0.009 | -0.1069 | No | ||
| 73 | UBE2R2 | 1230504 | 6951 | 0.009 | -0.1083 | No | ||
| 74 | ZBTB4 | 6450441 | 6973 | 0.009 | -0.1093 | No | ||
| 75 | ECT2 | 3830086 | 6975 | 0.009 | -0.1093 | No | ||
| 76 | MEF2C | 670025 780338 | 7171 | 0.008 | -0.1198 | No | ||
| 77 | OXR1 | 2370039 3190706 | 7416 | 0.007 | -0.1329 | No | ||
| 78 | RSBN1 | 7000487 | 7544 | 0.007 | -0.1397 | No | ||
| 79 | PLXNA1 | 70066 | 7596 | 0.007 | -0.1424 | No | ||
| 80 | WEE1 | 3390070 | 7696 | 0.006 | -0.1477 | No | ||
| 81 | ZKSCAN1 | 6380605 | 7815 | 0.006 | -0.1541 | No | ||
| 82 | OSBPL8 | 50603 | 7820 | 0.006 | -0.1542 | No | ||
| 83 | KCNMA1 | 1450402 | 7899 | 0.006 | -0.1584 | No | ||
| 84 | FOXL2 | 5670537 | 7946 | 0.005 | -0.1608 | No | ||
| 85 | LHX8 | 5700347 | 8124 | 0.005 | -0.1704 | No | ||
| 86 | TAOK2 | 110575 6380692 | 8258 | 0.004 | -0.1775 | No | ||
| 87 | PCDHA12 | 2810215 | 8264 | 0.004 | -0.1777 | No | ||
| 88 | VLDLR | 870722 3060047 5340452 6550131 | 8325 | 0.004 | -0.1809 | No | ||
| 89 | VSX1 | 4610605 6650112 | 8326 | 0.004 | -0.1809 | No | ||
| 90 | PCDHA10 | 3830725 | 8385 | 0.004 | -0.1840 | No | ||
| 91 | KLF12 | 1660095 4810288 5340546 6520286 | 8475 | 0.004 | -0.1888 | No | ||
| 92 | SLC2A4 | 540441 | 8545 | 0.004 | -0.1925 | No | ||
| 93 | SSX2IP | 4610176 | 8556 | 0.004 | -0.1930 | No | ||
| 94 | TLOC1 | 4050309 | 8825 | 0.003 | -0.2075 | No | ||
| 95 | PCDHA1 | 940026 540484 | 8963 | 0.002 | -0.2149 | No | ||
| 96 | TMEM16F | 5860008 | 8999 | 0.002 | -0.2168 | No | ||
| 97 | PRDM8 | 840746 | 9004 | 0.002 | -0.2170 | No | ||
| 98 | PCDHA3 | 3840402 | 9047 | 0.002 | -0.2192 | No | ||
| 99 | BRP44L | 1340605 | 9197 | 0.002 | -0.2273 | No | ||
| 100 | MMP24 | 5340154 | 9204 | 0.002 | -0.2276 | No | ||
| 101 | ANK2 | 6510546 | 9439 | 0.001 | -0.2403 | No | ||
| 102 | EPAS1 | 5290156 | 9554 | 0.001 | -0.2465 | No | ||
| 103 | ALX4 | 3870019 | 9569 | 0.001 | -0.2473 | No | ||
| 104 | ZBTB11 | 6520092 | 9578 | 0.001 | -0.2477 | No | ||
| 105 | BAMBI | 6650463 | 9589 | 0.001 | -0.2482 | No | ||
| 106 | OSBPL5 | 3840537 6040286 | 9654 | 0.000 | -0.2517 | No | ||
| 107 | PURA | 3870156 | 9881 | -0.000 | -0.2640 | No | ||
| 108 | ABCA1 | 6290156 | 9882 | -0.000 | -0.2640 | No | ||
| 109 | RAD23B | 2190671 | 10153 | -0.001 | -0.2787 | No | ||
| 110 | ZMYND11 | 630181 2570019 4850138 5360040 | 10251 | -0.001 | -0.2839 | No | ||
| 111 | GABPB2 | 6130450 7050735 | 10308 | -0.001 | -0.2869 | No | ||
| 112 | PCAF | 2230161 2570369 6550451 | 10437 | -0.002 | -0.2939 | No | ||
| 113 | PCDHA5 | 2970347 | 10716 | -0.003 | -0.3089 | No | ||
| 114 | PAK7 | 3830164 | 10833 | -0.003 | -0.3152 | No | ||
| 115 | NEUROD1 | 3060619 | 11107 | -0.004 | -0.3300 | No | ||
| 116 | MYT1L | 5670369 6660403 | 11303 | -0.005 | -0.3405 | No | ||
| 117 | ATAD2 | 870242 2260687 | 11466 | -0.005 | -0.3493 | No | ||
| 118 | NTN4 | 6940398 | 11487 | -0.005 | -0.3503 | No | ||
| 119 | HLF | 2370113 | 11494 | -0.005 | -0.3506 | No | ||
| 120 | CCNJ | 870047 | 11748 | -0.006 | -0.3642 | No | ||
| 121 | ATP2B2 | 3780397 | 12061 | -0.007 | -0.3811 | No | ||
| 122 | TRPV6 | 4010717 | 12068 | -0.007 | -0.3813 | No | ||
| 123 | PCDHA7 | 1850019 | 12292 | -0.008 | -0.3933 | No | ||
| 124 | LHX6 | 2360731 3360397 | 12331 | -0.009 | -0.3953 | No | ||
| 125 | E2F5 | 5860575 | 12458 | -0.009 | -0.4020 | No | ||
| 126 | NEUROD6 | 4670731 | 12460 | -0.009 | -0.4019 | No | ||
| 127 | CREB5 | 2320368 | 12683 | -0.010 | -0.4139 | No | ||
| 128 | PGBD5 | 1190047 | 12838 | -0.011 | -0.4221 | No | ||
| 129 | LUC7L2 | 1850402 6350088 | 12855 | -0.011 | -0.4228 | No | ||
| 130 | PAPOLA | 4200286 4850112 | 12858 | -0.011 | -0.4228 | No | ||
| 131 | SLITRK3 | 5910041 | 12921 | -0.012 | -0.4260 | No | ||
| 132 | PARP8 | 4150204 | 12978 | -0.012 | -0.4289 | No | ||
| 133 | NR4A3 | 2900021 5860095 5910039 | 12999 | -0.012 | -0.4298 | No | ||
| 134 | DHX40 | 360672 | 13171 | -0.014 | -0.4389 | No | ||
| 135 | PLEKHA3 | 5080068 | 13436 | -0.016 | -0.4531 | No | ||
| 136 | RB1CC1 | 510494 7100072 | 13448 | -0.016 | -0.4534 | No | ||
| 137 | PRRX1 | 4120193 4480390 | 13531 | -0.017 | -0.4577 | No | ||
| 138 | DDHD1 | 3450241 3940523 6380156 | 13620 | -0.018 | -0.4622 | No | ||
| 139 | CYP26B1 | 2630142 | 13635 | -0.018 | -0.4627 | No | ||
| 140 | UBE2B | 4780497 | 13779 | -0.020 | -0.4703 | No | ||
| 141 | GNB5 | 4480736 6550722 6770463 | 13943 | -0.022 | -0.4788 | No | ||
| 142 | TARDBP | 870390 2350093 | 13953 | -0.022 | -0.4790 | No | ||
| 143 | KLHL18 | 4560075 | 14042 | -0.023 | -0.4835 | No | ||
| 144 | ARID4A | 520156 1050707 | 14162 | -0.026 | -0.4896 | No | ||
| 145 | RGL1 | 1500097 6900450 | 14208 | -0.026 | -0.4917 | No | ||
| 146 | RAB11FIP1 | 2470333 | 14214 | -0.027 | -0.4916 | No | ||
| 147 | FGD5 | 3520438 4060500 | 14330 | -0.028 | -0.4975 | No | ||
| 148 | MUTED | 3140452 4610086 4730348 | 14457 | -0.031 | -0.5039 | No | ||
| 149 | MKNK2 | 1240020 5270092 | 14508 | -0.032 | -0.5062 | No | ||
| 150 | NR2C2 | 6100097 | 14667 | -0.037 | -0.5143 | No | ||
| 151 | ARHGEF17 | 2060008 | 14711 | -0.039 | -0.5162 | No | ||
| 152 | CFL2 | 380239 6200368 | 14783 | -0.041 | -0.5195 | No | ||
| 153 | MTUS1 | 780348 4920609 | 14792 | -0.042 | -0.5194 | No | ||
| 154 | PLAG1 | 1450142 3870139 6520039 | 14865 | -0.045 | -0.5227 | No | ||
| 155 | MINK1 | 5860647 | 14867 | -0.045 | -0.5221 | No | ||
| 156 | EPHA2 | 5890056 | 14973 | -0.050 | -0.5272 | No | ||
| 157 | CRIM1 | 130491 | 14979 | -0.051 | -0.5268 | No | ||
| 158 | YPEL2 | 5560609 | 15092 | -0.058 | -0.5321 | No | ||
| 159 | SNF1LK | 6110403 | 15238 | -0.067 | -0.5391 | No | ||
| 160 | BCL2L11 | 780044 4200601 | 15372 | -0.078 | -0.5453 | No | ||
| 161 | ZNF385 | 450176 4810358 | 15405 | -0.080 | -0.5459 | No | ||
| 162 | TAL1 | 7040239 | 15516 | -0.090 | -0.5507 | No | ||
| 163 | SUV39H1 | 630112 | 15532 | -0.092 | -0.5503 | No | ||
| 164 | MNT | 4920575 | 15554 | -0.096 | -0.5502 | No | ||
| 165 | RAB6A | 3840463 4780731 | 15611 | -0.102 | -0.5519 | No | ||
| 166 | E2F7 | 4200056 | 15660 | -0.106 | -0.5531 | No | ||
| 167 | DMTF1 | 5910047 1570497 2480100 6620239 | 15733 | -0.113 | -0.5555 | No | ||
| 168 | RPS6KA5 | 2120563 7040546 | 15771 | -0.117 | -0.5559 | No | ||
| 169 | HN1 | 3360156 | 15821 | -0.123 | -0.5570 | No | ||
| 170 | COL23A1 | 2030358 | 15995 | -0.142 | -0.5645 | No | ||
| 171 | FRMD4A | 2190176 | 16054 | -0.150 | -0.5656 | No | ||
| 172 | ORMDL3 | 1340711 | 16084 | -0.156 | -0.5651 | No | ||
| 173 | DNAJA2 | 2630253 6860358 | 16170 | -0.167 | -0.5675 | No | ||
| 174 | ATXN1 | 5550156 | 16338 | -0.192 | -0.5741 | No | ||
| 175 | RAB11A | 5360079 | 16383 | -0.201 | -0.5738 | No | ||
| 176 | CRK | 1230162 4780128 | 16406 | -0.206 | -0.5722 | No | ||
| 177 | AEBP2 | 2970603 | 16521 | -0.230 | -0.5754 | No | ||
| 178 | SPOP | 450035 | 16550 | -0.238 | -0.5737 | No | ||
| 179 | IQWD1 | 160475 | 16558 | -0.240 | -0.5709 | No | ||
| 180 | SMAD2 | 4200592 | 16670 | -0.263 | -0.5735 | No | ||
| 181 | PERQ1 | 1340164 | 16803 | -0.295 | -0.5767 | No | ||
| 182 | NFYA | 5860368 | 16825 | -0.301 | -0.5739 | No | ||
| 183 | MECP2 | 1940450 | 17044 | -0.360 | -0.5809 | Yes | ||
| 184 | BCL11A | 6860369 | 17052 | -0.364 | -0.5765 | Yes | ||
| 185 | HIC2 | 4010433 | 17104 | -0.385 | -0.5741 | Yes | ||
| 186 | SF4 | 3830341 | 17172 | -0.404 | -0.5724 | Yes | ||
| 187 | WDR37 | 2630519 6840347 | 17257 | -0.436 | -0.5712 | Yes | ||
| 188 | TUSC2 | 1580253 | 17392 | -0.492 | -0.5719 | Yes | ||
| 189 | MKRN1 | 1410097 | 17417 | -0.502 | -0.5665 | Yes | ||
| 190 | MAP3K14 | 5890435 | 17492 | -0.536 | -0.5634 | Yes | ||
| 191 | ZDHHC9 | 3940204 | 17505 | -0.541 | -0.5569 | Yes | ||
| 192 | MTMR3 | 1050014 4920180 | 17564 | -0.570 | -0.5525 | Yes | ||
| 193 | ASF1B | 6590706 | 17625 | -0.610 | -0.5476 | Yes | ||
| 194 | RRAGD | 670075 | 17646 | -0.622 | -0.5404 | Yes | ||
| 195 | ULK1 | 6100315 | 17720 | -0.669 | -0.5355 | Yes | ||
| 196 | LEF1 | 2470082 7100288 | 17770 | -0.695 | -0.5289 | Yes | ||
| 197 | CREBL1 | 1340546 | 17784 | -0.703 | -0.5203 | Yes | ||
| 198 | FOXJ2 | 2450441 | 17804 | -0.717 | -0.5118 | Yes | ||
| 199 | DUSP2 | 2370184 | 17814 | -0.722 | -0.5027 | Yes | ||
| 200 | MAP3K11 | 7000039 | 17815 | -0.725 | -0.4930 | Yes | ||
| 201 | RUNX1 | 3840711 | 17835 | -0.736 | -0.4843 | Yes | ||
| 202 | PPP6C | 2450670 | 17869 | -0.759 | -0.4760 | Yes | ||
| 203 | FEM1C | 60300 6130717 6980164 | 17876 | -0.765 | -0.4661 | Yes | ||
| 204 | TOB2 | 1240465 | 18083 | -0.970 | -0.4644 | Yes | ||
| 205 | IRF2 | 5390044 | 18100 | -0.987 | -0.4522 | Yes | ||
| 206 | WDR45 | 1660538 | 18101 | -0.988 | -0.4391 | Yes | ||
| 207 | CCND2 | 5340167 | 18184 | -1.101 | -0.4289 | Yes | ||
| 208 | ZFYVE26 | 6860152 | 18188 | -1.106 | -0.4143 | Yes | ||
| 209 | IHPK1 | 1340053 | 18237 | -1.184 | -0.4012 | Yes | ||
| 210 | KLF13 | 4670563 | 18251 | -1.209 | -0.3858 | Yes | ||
| 211 | MAML1 | 2760008 | 18267 | -1.263 | -0.3699 | Yes | ||
| 212 | MCL1 | 1660672 | 18315 | -1.362 | -0.3543 | Yes | ||
| 213 | PLAGL2 | 1770148 | 18356 | -1.489 | -0.3367 | Yes | ||
| 214 | TOX | 3440053 | 18388 | -1.613 | -0.3169 | Yes | ||
| 215 | ARHGEF18 | 2120279 | 18407 | -1.673 | -0.2956 | Yes | ||
| 216 | PLEKHM1 | 1050161 1990750 | 18412 | -1.692 | -0.2734 | Yes | ||
| 217 | PHF2 | 4060300 | 18421 | -1.715 | -0.2510 | Yes | ||
| 218 | MAPK9 | 2060273 3780209 4070397 | 18430 | -1.744 | -0.2283 | Yes | ||
| 219 | RASSF2 | 2480079 5720546 | 18520 | -2.347 | -0.2019 | Yes | ||
| 220 | TLE4 | 1450064 1500519 | 18558 | -2.750 | -0.1673 | Yes | ||
| 221 | BCL6 | 940100 | 18572 | -2.937 | -0.1290 | Yes | ||
| 222 | BTG1 | 4200735 6040131 6200133 | 18607 | -4.199 | -0.0750 | Yes | ||
| 223 | HRBL | 1850102 3830253 4780176 | 18611 | -5.676 | 0.0003 | Yes |

