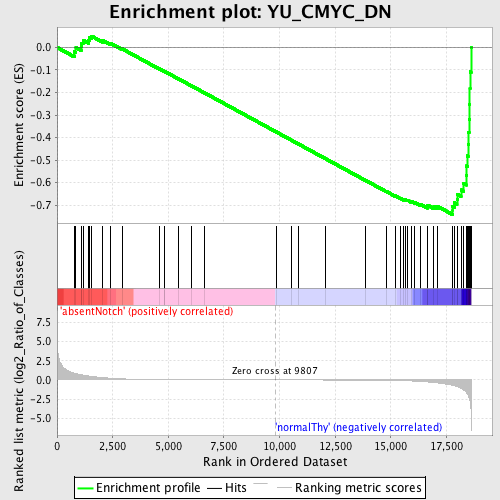
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | YU_CMYC_DN |
| Enrichment Score (ES) | -0.7417874 |
| Normalized Enrichment Score (NES) | -1.6137179 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.749 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IGH-1A | 360113 4670341 | 776 | 0.859 | -0.0192 | No | ||
| 2 | CTSH | 60524 | 847 | 0.814 | -0.0015 | No | ||
| 3 | UNC93B1 | 430152 | 1080 | 0.665 | 0.0035 | No | ||
| 4 | H2-DMB1 | 4760026 | 1101 | 0.654 | 0.0196 | No | ||
| 5 | H2-AB1 | 4230427 | 1169 | 0.622 | 0.0324 | No | ||
| 6 | H2-AA | 5290673 | 1420 | 0.505 | 0.0322 | No | ||
| 7 | GDI1 | 4590400 5290358 6020687 6180446 | 1464 | 0.488 | 0.0427 | No | ||
| 8 | PTPRC | 130402 5290148 | 1547 | 0.456 | 0.0503 | No | ||
| 9 | GNS | 3120458 | 2043 | 0.307 | 0.0317 | No | ||
| 10 | RBMS1 | 6400014 | 2400 | 0.222 | 0.0184 | No | ||
| 11 | H2-EB1 | 2350079 | 2920 | 0.136 | -0.0060 | No | ||
| 12 | OLFM1 | 3360161 5570066 | 4596 | 0.034 | -0.0953 | No | ||
| 13 | MACF1 | 1340132 2370176 2900750 6590364 | 4815 | 0.029 | -0.1063 | No | ||
| 14 | SRPK3 | 2480278 | 4831 | 0.029 | -0.1063 | No | ||
| 15 | GAD1 | 2360035 3140167 | 5465 | 0.019 | -0.1399 | No | ||
| 16 | PYGM | 3940300 | 6047 | 0.014 | -0.1708 | No | ||
| 17 | LOC207685 | 6520017 | 6610 | 0.011 | -0.2008 | No | ||
| 18 | ABCA1 | 6290156 | 9882 | -0.000 | -0.3770 | No | ||
| 19 | 5730420B22RIK | 130139 | 10528 | -0.002 | -0.4117 | No | ||
| 20 | SSPN | 4670091 | 10862 | -0.003 | -0.4296 | No | ||
| 21 | MYO7A | 2810102 | 12045 | -0.007 | -0.4930 | No | ||
| 22 | MS4A1 | 4210598 | 13841 | -0.021 | -0.5892 | No | ||
| 23 | GGA2 | 520451 1240082 2650332 | 14803 | -0.042 | -0.6398 | No | ||
| 24 | 1100001G20RIK | 3610368 | 15188 | -0.064 | -0.6588 | No | ||
| 25 | IFI203 | 6420102 3610369 | 15415 | -0.081 | -0.6689 | No | ||
| 26 | ACP5 | 2230717 | 15588 | -0.099 | -0.6755 | No | ||
| 27 | MGST1 | 6020605 | 15643 | -0.104 | -0.6757 | No | ||
| 28 | GBP4 | 6520471 1980670 | 15759 | -0.116 | -0.6788 | No | ||
| 29 | LAPTM5 | 5550372 | 15938 | -0.135 | -0.6849 | No | ||
| 30 | CMAH | 1660497 3390398 6550040 | 16066 | -0.153 | -0.6877 | No | ||
| 31 | BLK | 1940128 5390053 | 16340 | -0.193 | -0.6973 | No | ||
| 32 | H2-OB | 6980059 | 16638 | -0.256 | -0.7066 | No | ||
| 33 | H2-DMA | 6290463 | 16644 | -0.257 | -0.7000 | No | ||
| 34 | SIPA1 | 5220687 | 16935 | -0.330 | -0.7070 | No | ||
| 35 | PRKCB1 | 870019 3130092 | 17100 | -0.384 | -0.7057 | No | ||
| 36 | CD2 | 430672 | 17771 | -0.696 | -0.7235 | Yes | ||
| 37 | EVL | 1740113 | 17776 | -0.699 | -0.7053 | Yes | ||
| 38 | IL4I1 | 3190161 | 17850 | -0.746 | -0.6896 | Yes | ||
| 39 | RGL2 | 1740017 | 17975 | -0.845 | -0.6740 | Yes | ||
| 40 | CD37 | 770273 | 17996 | -0.869 | -0.6522 | Yes | ||
| 41 | CD1D1 | 5130184 | 18157 | -1.065 | -0.6328 | Yes | ||
| 42 | ARHGEF1 | 610347 4850603 6420672 | 18278 | -1.288 | -0.6053 | Yes | ||
| 43 | IL10RA | 1770164 | 18384 | -1.586 | -0.5692 | Yes | ||
| 44 | MAP4K2 | 4200037 | 18402 | -1.668 | -0.5262 | Yes | ||
| 45 | SLFN2 | 3120114 | 18443 | -1.777 | -0.4816 | Yes | ||
| 46 | ARID3B | 6650008 | 18476 | -1.964 | -0.4316 | Yes | ||
| 47 | DGKA | 5720152 5890328 | 18492 | -2.053 | -0.3784 | Yes | ||
| 48 | KLF2 | 6860270 | 18518 | -2.320 | -0.3187 | Yes | ||
| 49 | KLF3 | 5130438 | 18548 | -2.594 | -0.2519 | Yes | ||
| 50 | LTB | 3990672 | 18557 | -2.728 | -0.1805 | Yes | ||
| 51 | RAG1 | 2850279 | 18562 | -2.776 | -0.1077 | Yes | ||
| 52 | BTG1 | 4200735 6040131 6200133 | 18607 | -4.199 | 0.0005 | Yes |

