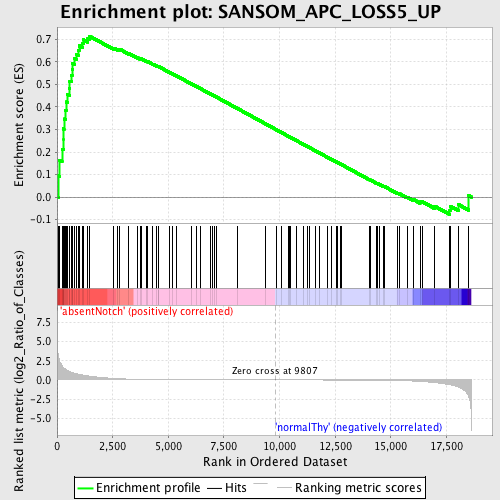
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | SANSOM_APC_LOSS5_UP |
| Enrichment Score (ES) | 0.7141429 |
| Normalized Enrichment Score (NES) | 1.5847371 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.10542534 |
| FWER p-Value | 0.763 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GNL3 | 4050338 4810500 | 49 | 3.279 | 0.0952 | Yes | ||
| 2 | NOL5 | 580731 | 117 | 2.416 | 0.1636 | Yes | ||
| 3 | EIF1 | 5080100 | 223 | 1.814 | 0.2121 | Yes | ||
| 4 | DDIT4 | 2570408 | 281 | 1.582 | 0.2562 | Yes | ||
| 5 | TXNL4A | 4120286 6510242 | 285 | 1.576 | 0.3031 | Yes | ||
| 6 | DDX21 | 6100446 | 313 | 1.509 | 0.3466 | Yes | ||
| 7 | HK2 | 2640722 | 384 | 1.365 | 0.3835 | Yes | ||
| 8 | NCL | 2360463 4540279 | 407 | 1.334 | 0.4221 | Yes | ||
| 9 | ETF1 | 6770075 | 472 | 1.218 | 0.4550 | Yes | ||
| 10 | CIRH1A | 2340372 | 550 | 1.097 | 0.4836 | Yes | ||
| 11 | NSUN2 | 2120193 | 570 | 1.071 | 0.5145 | Yes | ||
| 12 | PRKRIR | 2370402 | 640 | 0.994 | 0.5404 | Yes | ||
| 13 | GRWD1 | 6350528 | 676 | 0.946 | 0.5667 | Yes | ||
| 14 | GNL2 | 6620022 | 699 | 0.925 | 0.5931 | Yes | ||
| 15 | SLC27A1 | 6110093 | 770 | 0.867 | 0.6152 | Yes | ||
| 16 | COPS2 | 3060142 | 862 | 0.804 | 0.6343 | Yes | ||
| 17 | UTP14A | 6590048 | 959 | 0.744 | 0.6513 | Yes | ||
| 18 | FTSJ3 | 3140100 | 991 | 0.723 | 0.6712 | Yes | ||
| 19 | SMYD5 | 6220025 | 1137 | 0.639 | 0.6824 | Yes | ||
| 20 | PES1 | 2470427 | 1195 | 0.610 | 0.6975 | Yes | ||
| 21 | EIF4G2 | 3800575 6860184 | 1379 | 0.522 | 0.7032 | Yes | ||
| 22 | PRIM2A | 7000692 | 1452 | 0.495 | 0.7141 | Yes | ||
| 23 | CDCA7 | 3060097 | 2549 | 0.194 | 0.6608 | No | ||
| 24 | CDK9 | 630707 | 2725 | 0.165 | 0.6563 | No | ||
| 25 | FUSIP1 | 520082 5390114 | 2804 | 0.153 | 0.6566 | No | ||
| 26 | ADAM8 | 450347 5670609 | 3228 | 0.104 | 0.6369 | No | ||
| 27 | TNFRSF19 | 3870731 6940730 | 3626 | 0.074 | 0.6177 | No | ||
| 28 | RNF4 | 580286 1400152 | 3756 | 0.066 | 0.6127 | No | ||
| 29 | ORC2L | 1990470 6510019 | 3784 | 0.065 | 0.6132 | No | ||
| 30 | LAMC2 | 450692 2680041 4200059 4670148 | 4016 | 0.053 | 0.6023 | No | ||
| 31 | MYC | 380541 4670170 | 4075 | 0.051 | 0.6007 | No | ||
| 32 | CD24 | 1780091 | 4301 | 0.043 | 0.5898 | No | ||
| 33 | 2300002G24RIK | 2320040 | 4477 | 0.037 | 0.5815 | No | ||
| 34 | 9630050M13RIK | 2370086 | 4538 | 0.035 | 0.5793 | No | ||
| 35 | OLFR91 | 6290020 | 4544 | 0.035 | 0.5801 | No | ||
| 36 | SYNCRIP | 1690195 3140113 4670279 | 5035 | 0.025 | 0.5544 | No | ||
| 37 | ZRF1 | 6020181 6510484 | 5165 | 0.023 | 0.5481 | No | ||
| 38 | CEBPZ | 2970053 2680068 | 5355 | 0.021 | 0.5386 | No | ||
| 39 | EPHB3 | 6650341 | 6035 | 0.015 | 0.5024 | No | ||
| 40 | SEMA3C | 7000020 | 6245 | 0.013 | 0.4915 | No | ||
| 41 | PLA2G12A | 1740463 4010066 4050348 | 6441 | 0.012 | 0.4813 | No | ||
| 42 | KCND2 | 1170128 2100112 | 6883 | 0.009 | 0.4578 | No | ||
| 43 | PTBP2 | 1240398 | 6967 | 0.009 | 0.4536 | No | ||
| 44 | RIN2 | 2450184 | 7082 | 0.009 | 0.4477 | No | ||
| 45 | CITED1 | 2350670 | 7176 | 0.008 | 0.4429 | No | ||
| 46 | REPS1 | 2320603 2510333 | 8095 | 0.005 | 0.3935 | No | ||
| 47 | PLA2G2A | 3190458 | 9357 | 0.001 | 0.3256 | No | ||
| 48 | ZFP97 | 520324 | 9871 | -0.000 | 0.2979 | No | ||
| 49 | 1110014J01RIK | 1980332 | 10077 | -0.001 | 0.2868 | No | ||
| 50 | FOXA2 | 540338 5860441 | 10409 | -0.002 | 0.2690 | No | ||
| 51 | HELLS | 4560086 4810025 | 10446 | -0.002 | 0.2672 | No | ||
| 52 | ANP32B | 940095 2450494 4200368 | 10492 | -0.002 | 0.2648 | No | ||
| 53 | PKP1 | 1500433 | 10755 | -0.003 | 0.2507 | No | ||
| 54 | NUP54 | 940546 2360156 4060278 | 11083 | -0.004 | 0.2332 | No | ||
| 55 | KCNQ1 | 1580050 6860441 | 11275 | -0.004 | 0.2230 | No | ||
| 56 | FGFR4 | 2650072 | 11343 | -0.005 | 0.2195 | No | ||
| 57 | MMP7 | 1780497 | 11591 | -0.006 | 0.2064 | No | ||
| 58 | TSPAN12 | 4120010 | 11790 | -0.006 | 0.1959 | No | ||
| 59 | RNF149 | 5270494 | 12152 | -0.008 | 0.1766 | No | ||
| 60 | ZBTB17 | 6220082 | 12314 | -0.009 | 0.1682 | No | ||
| 61 | TGFBR1 | 1400148 4280020 6550711 | 12577 | -0.010 | 0.1544 | No | ||
| 62 | BTC | 3190133 | 12595 | -0.010 | 0.1538 | No | ||
| 63 | MYO1B | 770372 | 12743 | -0.011 | 0.1461 | No | ||
| 64 | FAM60A | 3940092 | 12782 | -0.011 | 0.1444 | No | ||
| 65 | 2810037C14RIK | 5290575 | 14053 | -0.024 | 0.0766 | No | ||
| 66 | KIF5A | 510435 | 14097 | -0.024 | 0.0750 | No | ||
| 67 | MDFI | 6200041 | 14358 | -0.029 | 0.0619 | No | ||
| 68 | CD44 | 3990072 4850671 5860411 6860148 7050551 | 14405 | -0.030 | 0.0603 | No | ||
| 69 | HDC | 4150463 | 14496 | -0.032 | 0.0564 | No | ||
| 70 | KIF20A | 2650050 | 14661 | -0.037 | 0.0486 | No | ||
| 71 | IFIT2 | 4810735 | 14706 | -0.038 | 0.0474 | No | ||
| 72 | HNRPAB | 540504 | 15300 | -0.072 | 0.0175 | No | ||
| 73 | ZZZ3 | 780133 2320239 2640452 5080706 6550341 | 15397 | -0.080 | 0.0147 | No | ||
| 74 | TIAM1 | 5420288 | 15730 | -0.113 | 0.0002 | No | ||
| 75 | SRR | 1190465 3800494 | 16016 | -0.145 | -0.0109 | No | ||
| 76 | MAP4K4 | 2360059 | 16313 | -0.188 | -0.0212 | No | ||
| 77 | ZFP90 | 580121 | 16404 | -0.205 | -0.0200 | No | ||
| 78 | FEM1B | 1230110 | 16966 | -0.339 | -0.0401 | No | ||
| 79 | 4930573I19RIK | 5270402 | 17652 | -0.624 | -0.0585 | No | ||
| 80 | SOX4 | 2260091 | 17671 | -0.637 | -0.0404 | No | ||
| 81 | 1190002H23RIK | 4480128 | 18037 | -0.912 | -0.0329 | No | ||
| 82 | ETS2 | 360451 | 18501 | -2.149 | 0.0062 | No |

