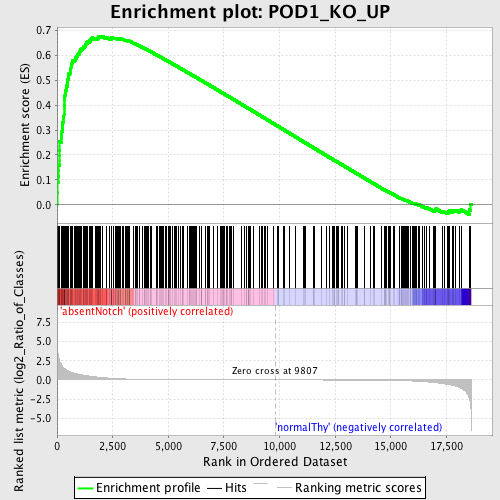
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | POD1_KO_UP |
| Enrichment Score (ES) | 0.67768425 |
| Normalized Enrichment Score (NES) | 1.7217994 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025319325 |
| FWER p-Value | 0.023 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PABPC1 | 2650180 2690253 6020632 1990270 | 3 | 6.314 | 0.0481 | Yes | ||
| 2 | PRDX2 | 6760095 4010619 5340577 | 8 | 5.586 | 0.0907 | Yes | ||
| 3 | PRR6 | 3450706 | 42 | 3.381 | 0.1147 | Yes | ||
| 4 | PCBP4 | 6350722 | 58 | 3.150 | 0.1380 | Yes | ||
| 5 | APEX1 | 3190519 | 66 | 3.039 | 0.1609 | Yes | ||
| 6 | GREB1 | 1230707 4560056 | 99 | 2.593 | 0.1790 | Yes | ||
| 7 | PSAT1 | 670446 1570102 | 102 | 2.559 | 0.1984 | Yes | ||
| 8 | ECM1 | 940068 5390309 | 105 | 2.534 | 0.2177 | Yes | ||
| 9 | PDLIM4 | 3990358 | 114 | 2.441 | 0.2359 | Yes | ||
| 10 | FCGRT | 5130315 | 115 | 2.429 | 0.2545 | Yes | ||
| 11 | BOK | 1170373 5860446 | 184 | 2.040 | 0.2664 | Yes | ||
| 12 | SRM | 1850097 2480162 | 192 | 2.000 | 0.2813 | Yes | ||
| 13 | SALL2 | 2190372 | 216 | 1.852 | 0.2942 | Yes | ||
| 14 | TMEM132A | 510433 770711 | 230 | 1.749 | 0.3069 | Yes | ||
| 15 | THOP1 | 2350450 | 238 | 1.722 | 0.3197 | Yes | ||
| 16 | LGALS1 | 6450193 | 248 | 1.683 | 0.3321 | Yes | ||
| 17 | TGFBI | 2060446 6900112 | 291 | 1.548 | 0.3416 | Yes | ||
| 18 | GJA1 | 5220731 | 295 | 1.543 | 0.3533 | Yes | ||
| 19 | PYCRL | 4780204 | 309 | 1.518 | 0.3642 | Yes | ||
| 20 | GUK1 | 5900059 | 322 | 1.483 | 0.3749 | Yes | ||
| 21 | TGM2 | 5360452 | 323 | 1.483 | 0.3862 | Yes | ||
| 22 | ACTB | 1660736 2030204 2060215 5050047 | 326 | 1.476 | 0.3974 | Yes | ||
| 23 | PPAN | 540398 | 333 | 1.469 | 0.4083 | Yes | ||
| 24 | CHAF1B | 3940142 | 334 | 1.467 | 0.4195 | Yes | ||
| 25 | GALK1 | 840162 | 340 | 1.455 | 0.4304 | Yes | ||
| 26 | CELSR1 | 6510348 | 350 | 1.434 | 0.4409 | Yes | ||
| 27 | CAD | 2340717 | 375 | 1.384 | 0.4502 | Yes | ||
| 28 | TBRG4 | 3610239 6370594 | 383 | 1.368 | 0.4603 | Yes | ||
| 29 | PHGDH | 1500280 | 410 | 1.325 | 0.4690 | Yes | ||
| 30 | SUMO3 | 130600 3710136 | 435 | 1.290 | 0.4775 | Yes | ||
| 31 | APRT | 7000403 | 455 | 1.246 | 0.4860 | Yes | ||
| 32 | ABHD14A | 4670670 | 460 | 1.233 | 0.4952 | Yes | ||
| 33 | SIPA1L1 | 50292 | 476 | 1.207 | 0.5037 | Yes | ||
| 34 | TIMM13 | 2450601 | 501 | 1.158 | 0.5112 | Yes | ||
| 35 | ANTXR1 | 1990593 2850056 | 523 | 1.129 | 0.5187 | Yes | ||
| 36 | NLGN2 | 5900592 | 530 | 1.115 | 0.5269 | Yes | ||
| 37 | CLU | 5420075 | 580 | 1.061 | 0.5323 | Yes | ||
| 38 | MIF | 3710332 | 587 | 1.052 | 0.5400 | Yes | ||
| 39 | STARD10 | 1400619 4570170 | 599 | 1.041 | 0.5474 | Yes | ||
| 40 | MNS1 | 5690528 | 624 | 1.015 | 0.5539 | Yes | ||
| 41 | PPP1R14B | 3800075 | 643 | 0.990 | 0.5605 | Yes | ||
| 42 | EXOSC5 | 3370671 | 659 | 0.963 | 0.5670 | Yes | ||
| 43 | GRWD1 | 6350528 | 676 | 0.946 | 0.5734 | Yes | ||
| 44 | CYP1B1 | 4760735 | 703 | 0.923 | 0.5790 | Yes | ||
| 45 | COX4NB | 1980044 3610014 | 793 | 0.843 | 0.5806 | Yes | ||
| 46 | SNRP70 | 4200181 | 815 | 0.832 | 0.5858 | Yes | ||
| 47 | PA2G4 | 2030463 | 823 | 0.827 | 0.5918 | Yes | ||
| 48 | C16ORF61 | 6290575 6760403 | 864 | 0.804 | 0.5957 | Yes | ||
| 49 | POLD2 | 6400148 | 900 | 0.778 | 0.5998 | Yes | ||
| 50 | PFKL | 6200167 | 934 | 0.759 | 0.6038 | Yes | ||
| 51 | FKBP4 | 870035 | 954 | 0.748 | 0.6084 | Yes | ||
| 52 | MGP | 6900736 | 1005 | 0.712 | 0.6112 | Yes | ||
| 53 | UNC119 | 360372 | 1007 | 0.709 | 0.6165 | Yes | ||
| 54 | LSM3 | 780164 | 1044 | 0.688 | 0.6198 | Yes | ||
| 55 | RAB17 | 6110019 6220619 | 1057 | 0.677 | 0.6243 | Yes | ||
| 56 | RFC3 | 1980600 | 1114 | 0.649 | 0.6262 | Yes | ||
| 57 | EVC2 | 5700440 | 1163 | 0.628 | 0.6284 | Yes | ||
| 58 | VCAM1 | 2900450 | 1185 | 0.613 | 0.6320 | Yes | ||
| 59 | ABHD11 | 1240086 | 1207 | 0.602 | 0.6354 | Yes | ||
| 60 | POLE3 | 5270019 | 1248 | 0.579 | 0.6377 | Yes | ||
| 61 | NFIX | 2450152 | 1286 | 0.564 | 0.6400 | Yes | ||
| 62 | STMN1 | 1990717 | 1289 | 0.561 | 0.6441 | Yes | ||
| 63 | EXOSC8 | 360427 | 1302 | 0.557 | 0.6478 | Yes | ||
| 64 | WISP1 | 2340019 3130692 | 1303 | 0.557 | 0.6520 | Yes | ||
| 65 | BICC1 | 4670647 | 1357 | 0.527 | 0.6532 | Yes | ||
| 66 | FKBP10 | 1340706 | 1365 | 0.525 | 0.6568 | Yes | ||
| 67 | STRA13 | 2650039 6900373 | 1451 | 0.496 | 0.6559 | Yes | ||
| 68 | ETV5 | 110017 | 1455 | 0.493 | 0.6595 | Yes | ||
| 69 | LSM4 | 3130180 | 1496 | 0.479 | 0.6610 | Yes | ||
| 70 | PYCR1 | 6180670 | 1515 | 0.469 | 0.6636 | Yes | ||
| 71 | LMNB1 | 5890292 6020008 | 1526 | 0.464 | 0.6666 | Yes | ||
| 72 | SOAT1 | 1190019 | 1557 | 0.451 | 0.6684 | Yes | ||
| 73 | MCM3 | 5570068 | 1578 | 0.444 | 0.6708 | Yes | ||
| 74 | PRIM1 | 6420746 | 1709 | 0.403 | 0.6667 | Yes | ||
| 75 | CXXC6 | 2810168 | 1791 | 0.380 | 0.6652 | Yes | ||
| 76 | SLC38A1 | 520465 | 1801 | 0.374 | 0.6676 | Yes | ||
| 77 | MRPS27 | 4850546 | 1807 | 0.372 | 0.6702 | Yes | ||
| 78 | GAS6 | 4480021 | 1847 | 0.360 | 0.6708 | Yes | ||
| 79 | SERTAD4 | 130593 | 1850 | 0.359 | 0.6734 | Yes | ||
| 80 | NKD1 | 1340286 | 1854 | 0.358 | 0.6760 | Yes | ||
| 81 | BAT5 | 5720687 | 1902 | 0.346 | 0.6761 | Yes | ||
| 82 | RHEBL1 | 2810576 | 1959 | 0.327 | 0.6755 | Yes | ||
| 83 | PPIH | 5910403 | 1966 | 0.326 | 0.6777 | Yes | ||
| 84 | TPX2 | 6420324 | 2061 | 0.302 | 0.6749 | No | ||
| 85 | IGSF8 | 1580592 | 2205 | 0.264 | 0.6691 | No | ||
| 86 | RAD18 | 5670372 | 2236 | 0.256 | 0.6694 | No | ||
| 87 | CNDP2 | 6590333 | 2237 | 0.256 | 0.6713 | No | ||
| 88 | VPS72 | 2450181 | 2356 | 0.231 | 0.6667 | No | ||
| 89 | LRFN4 | 5670368 | 2422 | 0.219 | 0.6648 | No | ||
| 90 | MCM5 | 2680647 | 2428 | 0.218 | 0.6662 | No | ||
| 91 | NEURL | 5390538 | 2432 | 0.217 | 0.6677 | No | ||
| 92 | RBP1 | 1690072 | 2440 | 0.216 | 0.6690 | No | ||
| 93 | TRPS1 | 870100 | 2453 | 0.213 | 0.6699 | No | ||
| 94 | ASPM | 3390156 | 2460 | 0.212 | 0.6712 | No | ||
| 95 | TGFB3 | 1070041 | 2515 | 0.200 | 0.6698 | No | ||
| 96 | CDCA7 | 3060097 | 2549 | 0.194 | 0.6695 | No | ||
| 97 | RELN | 6510037 | 2609 | 0.184 | 0.6677 | No | ||
| 98 | TAF6L | 1850056 | 2630 | 0.179 | 0.6680 | No | ||
| 99 | NCAPH | 6220435 | 2651 | 0.177 | 0.6682 | No | ||
| 100 | MCM7 | 3290292 5220056 | 2697 | 0.169 | 0.6670 | No | ||
| 101 | CDKN1B | 3800025 6450044 | 2706 | 0.167 | 0.6679 | No | ||
| 102 | NUP98 | 2850465 | 2748 | 0.161 | 0.6669 | No | ||
| 103 | COL6A1 | 1410670 2940091 6510086 | 2760 | 0.160 | 0.6675 | No | ||
| 104 | DTYMK | 2340377 | 2768 | 0.159 | 0.6683 | No | ||
| 105 | IGF1 | 1990193 3130377 3290280 | 2798 | 0.153 | 0.6679 | No | ||
| 106 | MRPL9 | 580079 | 2837 | 0.149 | 0.6670 | No | ||
| 107 | SRD5A1 | 5910347 | 2858 | 0.145 | 0.6670 | No | ||
| 108 | CDCA5 | 5670131 | 2864 | 0.145 | 0.6678 | No | ||
| 109 | BZW2 | 940079 | 2925 | 0.136 | 0.6656 | No | ||
| 110 | KNTC1 | 430079 | 2962 | 0.132 | 0.6646 | No | ||
| 111 | PAPSS2 | 870113 | 3069 | 0.121 | 0.6598 | No | ||
| 112 | POLD1 | 4830026 | 3082 | 0.120 | 0.6601 | No | ||
| 113 | WDR34 | 6100215 | 3089 | 0.119 | 0.6606 | No | ||
| 114 | VPS54 | 1660168 | 3112 | 0.116 | 0.6603 | No | ||
| 115 | CPXM1 | 5390725 | 3141 | 0.113 | 0.6597 | No | ||
| 116 | HNRPU | 6520736 | 3175 | 0.110 | 0.6587 | No | ||
| 117 | TK1 | 540014 540494 6760152 | 3189 | 0.109 | 0.6588 | No | ||
| 118 | CYBA | 4760739 | 3232 | 0.104 | 0.6573 | No | ||
| 119 | B4GALT2 | 780037 | 3257 | 0.101 | 0.6568 | No | ||
| 120 | NDUFB7 | 1850026 | 3262 | 0.101 | 0.6573 | No | ||
| 121 | SPSB4 | 4070673 | 3263 | 0.101 | 0.6581 | No | ||
| 122 | YPEL1 | 6590435 | 3441 | 0.086 | 0.6491 | No | ||
| 123 | EPHB2 | 1090288 1410377 2100541 | 3504 | 0.082 | 0.6463 | No | ||
| 124 | TNFRSF19L | 1170168 1190358 5670270 | 3517 | 0.081 | 0.6463 | No | ||
| 125 | XRCC1 | 4210609 | 3565 | 0.078 | 0.6443 | No | ||
| 126 | DCN | 510332 5340026 5900711 6550092 | 3621 | 0.075 | 0.6419 | No | ||
| 127 | SERPINF1 | 7040367 | 3682 | 0.071 | 0.6391 | No | ||
| 128 | LGI2 | 520056 | 3704 | 0.069 | 0.6385 | No | ||
| 129 | CACNB2 | 1500095 7330707 | 3848 | 0.061 | 0.6312 | No | ||
| 130 | SKP2 | 360711 380093 4810368 | 3915 | 0.058 | 0.6280 | No | ||
| 131 | SGTA | 3940154 | 3925 | 0.057 | 0.6280 | No | ||
| 132 | LSR | 510647 1400402 | 3956 | 0.056 | 0.6268 | No | ||
| 133 | SNRPB | 1230097 2940086 3520671 | 4027 | 0.053 | 0.6233 | No | ||
| 134 | CDT1 | 3780682 | 4051 | 0.051 | 0.6225 | No | ||
| 135 | MAPKAPK3 | 630520 | 4093 | 0.050 | 0.6206 | No | ||
| 136 | CA4 | 130100 | 4183 | 0.046 | 0.6161 | No | ||
| 137 | PLTP | 630110 130497 4050139 | 4193 | 0.046 | 0.6160 | No | ||
| 138 | COL3A1 | 6420273 6620044 | 4235 | 0.045 | 0.6141 | No | ||
| 139 | CPNE5 | 6380673 | 4457 | 0.038 | 0.6023 | No | ||
| 140 | SLC4A3 | 360603 4730239 6380347 | 4473 | 0.037 | 0.6018 | No | ||
| 141 | SOX11 | 610279 | 4515 | 0.036 | 0.5998 | No | ||
| 142 | EDNRA | 6900133 | 4616 | 0.033 | 0.5946 | No | ||
| 143 | ESR1 | 4060372 5860193 | 4638 | 0.033 | 0.5937 | No | ||
| 144 | SESN2 | 2370019 | 4670 | 0.032 | 0.5922 | No | ||
| 145 | MMP11 | 1980619 | 4747 | 0.030 | 0.5883 | No | ||
| 146 | FZD3 | 1500397 | 4788 | 0.029 | 0.5864 | No | ||
| 147 | RHOU | 2470706 | 4883 | 0.028 | 0.5814 | No | ||
| 148 | SIX2 | 1450551 | 4931 | 0.027 | 0.5791 | No | ||
| 149 | HMMR | 5720315 6380168 | 4989 | 0.026 | 0.5762 | No | ||
| 150 | SAP18 | 3610092 | 5003 | 0.026 | 0.5756 | No | ||
| 151 | DDEF1 | 1170411 4070465 | 5020 | 0.025 | 0.5750 | No | ||
| 152 | NCAM1 | 3140026 | 5032 | 0.025 | 0.5746 | No | ||
| 153 | METTL1 | 4540670 5860162 7040433 | 5099 | 0.024 | 0.5711 | No | ||
| 154 | TNPO1 | 730092 2260735 | 5176 | 0.023 | 0.5672 | No | ||
| 155 | CDC25C | 2570673 4760161 6520707 | 5258 | 0.022 | 0.5629 | No | ||
| 156 | FLNC | 2360048 5050270 | 5329 | 0.021 | 0.5592 | No | ||
| 157 | SASH1 | 4070270 | 5346 | 0.021 | 0.5585 | No | ||
| 158 | PTGDS | 3610519 | 5440 | 0.020 | 0.5536 | No | ||
| 159 | LBP | 6860019 | 5567 | 0.018 | 0.5468 | No | ||
| 160 | CCNA2 | 5290075 | 5631 | 0.018 | 0.5435 | No | ||
| 161 | SNAP91 | 3170056 5690520 | 5638 | 0.018 | 0.5434 | No | ||
| 162 | SRPX | 580717 1980162 | 5666 | 0.017 | 0.5420 | No | ||
| 163 | FN1 | 1170601 2970647 6220288 6940037 | 5872 | 0.016 | 0.5309 | No | ||
| 164 | ZFPM2 | 460072 | 5951 | 0.015 | 0.5268 | No | ||
| 165 | PTN | 5910161 | 5967 | 0.015 | 0.5261 | No | ||
| 166 | TEAD2 | 6450079 | 5982 | 0.015 | 0.5254 | No | ||
| 167 | HNRPR | 2320440 2900601 4610008 | 6049 | 0.014 | 0.5219 | No | ||
| 168 | ALCAM | 1050019 | 6077 | 0.014 | 0.5206 | No | ||
| 169 | PTP4A1 | 130692 | 6117 | 0.014 | 0.5185 | No | ||
| 170 | COL6A2 | 1780142 | 6153 | 0.014 | 0.5167 | No | ||
| 171 | NFIA | 2760129 5860278 | 6211 | 0.013 | 0.5137 | No | ||
| 172 | SEMA3C | 7000020 | 6245 | 0.013 | 0.5120 | No | ||
| 173 | NFATC4 | 2470735 | 6252 | 0.013 | 0.5118 | No | ||
| 174 | CDH1 | 1940736 | 6404 | 0.012 | 0.5036 | No | ||
| 175 | WNT4 | 4150619 | 6493 | 0.012 | 0.4989 | No | ||
| 176 | CDH11 | 1230133 2100180 4610110 | 6500 | 0.012 | 0.4987 | No | ||
| 177 | BLM | 520619 5570170 | 6503 | 0.012 | 0.4987 | No | ||
| 178 | DSTN | 3140592 | 6654 | 0.011 | 0.4905 | No | ||
| 179 | CYP7B1 | 1190309 | 6687 | 0.011 | 0.4889 | No | ||
| 180 | PAX8 | 5050301 5290164 | 6754 | 0.010 | 0.4854 | No | ||
| 181 | PRDX6 | 4920397 6380601 | 6776 | 0.010 | 0.4843 | No | ||
| 182 | PHACTR1 | 3190176 5670286 | 6793 | 0.010 | 0.4835 | No | ||
| 183 | TRPM4 | 6980270 | 6837 | 0.010 | 0.4812 | No | ||
| 184 | BMPER | 4780520 | 7042 | 0.009 | 0.4701 | No | ||
| 185 | MTMR7 | 1170450 | 7203 | 0.008 | 0.4615 | No | ||
| 186 | GPC6 | 5270162 | 7340 | 0.008 | 0.4541 | No | ||
| 187 | FRAS1 | 5080170 5670347 | 7383 | 0.007 | 0.4518 | No | ||
| 188 | ATXN7 | 1450435 | 7411 | 0.007 | 0.4504 | No | ||
| 189 | CDH6 | 4730541 | 7480 | 0.007 | 0.4468 | No | ||
| 190 | EYA1 | 1450278 5220390 | 7489 | 0.007 | 0.4464 | No | ||
| 191 | LAMA2 | 7040020 | 7539 | 0.007 | 0.4438 | No | ||
| 192 | FMOD | 540577 | 7604 | 0.007 | 0.4403 | No | ||
| 193 | HUNK | 7100139 | 7608 | 0.007 | 0.4402 | No | ||
| 194 | MFAP2 | 2970685 3060300 | 7644 | 0.007 | 0.4383 | No | ||
| 195 | PCDH9 | 2100136 | 7761 | 0.006 | 0.4320 | No | ||
| 196 | CTHRC1 | 4150020 | 7796 | 0.006 | 0.4302 | No | ||
| 197 | VPS37D | 3190142 | 7830 | 0.006 | 0.4285 | No | ||
| 198 | BNC2 | 4810603 | 7836 | 0.006 | 0.4282 | No | ||
| 199 | FNDC4 | 3870440 5860576 | 7913 | 0.006 | 0.4241 | No | ||
| 200 | RGS17 | 1400603 | 8309 | 0.004 | 0.4026 | No | ||
| 201 | CCNB2 | 6510528 | 8400 | 0.004 | 0.3977 | No | ||
| 202 | NOPE | 380156 | 8505 | 0.004 | 0.3921 | No | ||
| 203 | COL8A2 | 430020 | 8531 | 0.004 | 0.3907 | No | ||
| 204 | SLIT2 | 1940037 | 8580 | 0.004 | 0.3881 | No | ||
| 205 | SPON1 | 1660301 | 8584 | 0.004 | 0.3880 | No | ||
| 206 | DNAJC9 | 6130019 | 8608 | 0.003 | 0.3868 | No | ||
| 207 | NIPSNAP1 | 6860138 | 8654 | 0.003 | 0.3843 | No | ||
| 208 | ARSB | 3190093 5360491 | 8674 | 0.003 | 0.3833 | No | ||
| 209 | PCDH8 | 2360047 4200452 | 8829 | 0.003 | 0.3749 | No | ||
| 210 | FAP | 4560019 | 9115 | 0.002 | 0.3594 | No | ||
| 211 | TNFAIP6 | 3390161 | 9190 | 0.002 | 0.3553 | No | ||
| 212 | RCN3 | 5420400 | 9252 | 0.002 | 0.3520 | No | ||
| 213 | MATN2 | 460215 | 9315 | 0.001 | 0.3486 | No | ||
| 214 | MAOA | 1410039 4610324 | 9368 | 0.001 | 0.3458 | No | ||
| 215 | ADCY2 | 4610100 | 9447 | 0.001 | 0.3416 | No | ||
| 216 | PHB | 2260739 | 9723 | 0.000 | 0.3265 | No | ||
| 217 | NRP2 | 4070400 5860041 6650446 | 9922 | -0.000 | 0.3157 | No | ||
| 218 | HOMER2 | 840242 | 9931 | -0.000 | 0.3153 | No | ||
| 219 | HMGB3 | 2940168 | 9953 | -0.000 | 0.3141 | No | ||
| 220 | CFTR | 1850008 | 10154 | -0.001 | 0.3032 | No | ||
| 221 | TIMELESS | 3710315 | 10212 | -0.001 | 0.3001 | No | ||
| 222 | HELLS | 4560086 4810025 | 10446 | -0.002 | 0.2874 | No | ||
| 223 | LUM | 5420079 | 10703 | -0.003 | 0.2734 | No | ||
| 224 | BMP4 | 380113 | 11085 | -0.004 | 0.2527 | No | ||
| 225 | KCND3 | 670041 | 11113 | -0.004 | 0.2512 | No | ||
| 226 | CLSTN2 | 6900546 | 11158 | -0.004 | 0.2488 | No | ||
| 227 | BOC | 5270348 | 11505 | -0.005 | 0.2300 | No | ||
| 228 | RAD51 | 6110450 6980280 | 11516 | -0.005 | 0.2295 | No | ||
| 229 | EFS | 110288 | 11559 | -0.005 | 0.2272 | No | ||
| 230 | BMP5 | 4670048 | 11864 | -0.007 | 0.2107 | No | ||
| 231 | GATA6 | 1770010 | 12097 | -0.008 | 0.1980 | No | ||
| 232 | HS6ST2 | 520133 5550603 | 12227 | -0.008 | 0.1911 | No | ||
| 233 | LHFPL2 | 450020 | 12359 | -0.009 | 0.1840 | No | ||
| 234 | FBLN5 | 6550010 | 12427 | -0.009 | 0.1804 | No | ||
| 235 | CAPN6 | 1740168 | 12459 | -0.009 | 0.1788 | No | ||
| 236 | C1QL1 | 2030465 | 12463 | -0.009 | 0.1787 | No | ||
| 237 | TCF2 | 870338 5050632 | 12543 | -0.010 | 0.1744 | No | ||
| 238 | HOXB5 | 2900538 | 12615 | -0.010 | 0.1706 | No | ||
| 239 | MEIS1 | 1400575 | 12634 | -0.010 | 0.1697 | No | ||
| 240 | POSTN | 450411 6040451 | 12661 | -0.010 | 0.1684 | No | ||
| 241 | SPINT1 | 3190059 | 12772 | -0.011 | 0.1625 | No | ||
| 242 | DCAMKL1 | 540095 2690092 | 12820 | -0.011 | 0.1600 | No | ||
| 243 | PDZRN3 | 2340131 | 12912 | -0.012 | 0.1551 | No | ||
| 244 | PIN1 | 2970408 | 12925 | -0.012 | 0.1545 | No | ||
| 245 | HMGA2 | 2940121 3390647 5130279 6400136 | 13058 | -0.013 | 0.1474 | No | ||
| 246 | ACE2 | 3360446 | 13391 | -0.015 | 0.1294 | No | ||
| 247 | IGFBP2 | 5220563 | 13443 | -0.016 | 0.1267 | No | ||
| 248 | CBX5 | 3830072 6290167 | 13476 | -0.016 | 0.1251 | No | ||
| 249 | GLRB | 2510433 4280736 | 13500 | -0.016 | 0.1240 | No | ||
| 250 | RPL41 | 6940112 | 13805 | -0.020 | 0.1075 | No | ||
| 251 | PTPRK | 1450433 2640142 | 13831 | -0.020 | 0.1063 | No | ||
| 252 | CFH | 1240053 2030541 5900731 | 14076 | -0.024 | 0.0932 | No | ||
| 253 | GRIP1 | 380280 6450435 6860632 | 14209 | -0.026 | 0.0862 | No | ||
| 254 | FRAP1 | 2850037 6660010 | 14253 | -0.027 | 0.0840 | No | ||
| 255 | TMEPAI | 1340253 | 14599 | -0.035 | 0.0654 | No | ||
| 256 | PTPN12 | 2030309 6020725 6130746 6290609 | 14704 | -0.038 | 0.0600 | No | ||
| 257 | LTBP1 | 780035 2100746 2760402 3780551 6550184 6860193 | 14737 | -0.040 | 0.0586 | No | ||
| 258 | TTL | 6220048 | 14776 | -0.041 | 0.0568 | No | ||
| 259 | AGTR2 | 2450632 | 14781 | -0.041 | 0.0569 | No | ||
| 260 | GRIA3 | 2900164 6130278 6860154 | 14816 | -0.043 | 0.0554 | No | ||
| 261 | COMP | 3800273 | 14875 | -0.045 | 0.0526 | No | ||
| 262 | MDK | 3840020 | 14925 | -0.048 | 0.0503 | No | ||
| 263 | EPHA4 | 460750 | 14940 | -0.049 | 0.0499 | No | ||
| 264 | KCNK5 | 4590504 6860687 | 14982 | -0.051 | 0.0480 | No | ||
| 265 | SMC1A | 3060600 5700148 5890113 6370154 | 15141 | -0.061 | 0.0399 | No | ||
| 266 | PALM | 130601 3850017 | 15155 | -0.061 | 0.0396 | No | ||
| 267 | LAMA4 | 3170647 | 15377 | -0.079 | 0.0282 | No | ||
| 268 | NPHP1 | 4480332 4560286 | 15409 | -0.081 | 0.0271 | No | ||
| 269 | BANF1 | 460164 2850170 6110139 | 15465 | -0.086 | 0.0247 | No | ||
| 270 | APBB1 | 2690338 | 15479 | -0.087 | 0.0247 | No | ||
| 271 | SUV39H1 | 630112 | 15532 | -0.092 | 0.0225 | No | ||
| 272 | CDCA3 | 3940397 | 15585 | -0.099 | 0.0205 | No | ||
| 273 | TACC3 | 5130592 | 15617 | -0.102 | 0.0196 | No | ||
| 274 | IPO13 | 3870088 | 15655 | -0.106 | 0.0183 | No | ||
| 275 | CLEC11A | 2680193 | 15711 | -0.111 | 0.0162 | No | ||
| 276 | TIAM1 | 5420288 | 15730 | -0.113 | 0.0161 | No | ||
| 277 | PDGFA | 6020095 | 15778 | -0.118 | 0.0144 | No | ||
| 278 | KLF5 | 3840348 | 15876 | -0.129 | 0.0101 | No | ||
| 279 | CDCA8 | 2340286 6980019 | 15885 | -0.130 | 0.0106 | No | ||
| 280 | PTTG1 | 520463 | 15985 | -0.141 | 0.0063 | No | ||
| 281 | MORN2 | 6760333 | 16035 | -0.148 | 0.0048 | No | ||
| 282 | SSSCA1 | 1240402 780309 | 16056 | -0.150 | 0.0048 | No | ||
| 283 | FXN | 4070500 | 16115 | -0.160 | 0.0029 | No | ||
| 284 | CLCN2 | 1940021 | 16149 | -0.164 | 0.0023 | No | ||
| 285 | BRUNOL4 | 4490452 5080451 | 16157 | -0.165 | 0.0032 | No | ||
| 286 | CENPK | 1740722 6200068 | 16255 | -0.179 | -0.0007 | No | ||
| 287 | MSI2 | 1190066 5270039 | 16271 | -0.180 | -0.0002 | No | ||
| 288 | ATIC | 4010593 6380463 | 16435 | -0.211 | -0.0074 | No | ||
| 289 | EMG1 | 50286 380242 | 16527 | -0.231 | -0.0107 | No | ||
| 290 | PAFAH1B3 | 3120168 | 16587 | -0.245 | -0.0120 | No | ||
| 291 | UBE2C | 6130017 | 16589 | -0.245 | -0.0102 | No | ||
| 292 | KIF22 | 1190368 | 16735 | -0.278 | -0.0160 | No | ||
| 293 | FRAG1 | 1050504 1170152 2030092 | 16937 | -0.330 | -0.0244 | No | ||
| 294 | SBK1 | 940452 | 16963 | -0.337 | -0.0232 | No | ||
| 295 | ADD3 | 4150739 5390600 | 16993 | -0.347 | -0.0221 | No | ||
| 296 | CDC45L | 70537 3130114 | 17001 | -0.348 | -0.0199 | No | ||
| 297 | ADIPOR2 | 1510035 | 17005 | -0.349 | -0.0174 | No | ||
| 298 | SOCS2 | 4760692 | 17029 | -0.355 | -0.0159 | No | ||
| 299 | CENPA | 5080154 | 17309 | -0.458 | -0.0276 | No | ||
| 300 | RANBP1 | 430215 1090180 | 17406 | -0.499 | -0.0291 | No | ||
| 301 | BCL7A | 4560095 | 17550 | -0.562 | -0.0326 | No | ||
| 302 | INCENP | 520593 | 17574 | -0.576 | -0.0294 | No | ||
| 303 | PPP2CA | 3990113 | 17601 | -0.596 | -0.0263 | No | ||
| 304 | BEX2 | 5670433 | 17644 | -0.618 | -0.0238 | No | ||
| 305 | SEPW1 | 3800079 | 17762 | -0.690 | -0.0250 | No | ||
| 306 | UHRF1 | 1410273 1660242 | 17828 | -0.731 | -0.0229 | No | ||
| 307 | BACH2 | 6760131 | 17928 | -0.809 | -0.0221 | No | ||
| 308 | TYMS | 940450 1940068 3710008 5570546 | 18068 | -0.958 | -0.0224 | No | ||
| 309 | CCND2 | 5340167 | 18184 | -1.101 | -0.0203 | No | ||
| 310 | CD79B | 1450066 3390358 | 18529 | -2.449 | -0.0203 | No | ||
| 311 | LSP1 | 2350091 | 18586 | -3.268 | 0.0016 | No |

