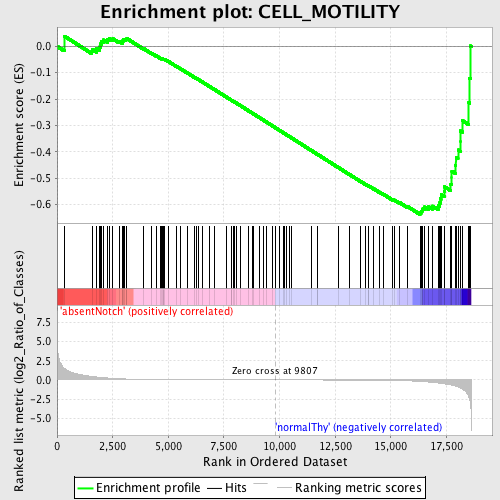
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | CELL_MOTILITY |
| Enrichment Score (ES) | -0.63795817 |
| Normalized Enrichment Score (NES) | -1.528537 |
| Nominal p-value | 0.0026041667 |
| FDR q-value | 0.41354707 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ACTB | 1660736 2030204 2060215 5050047 | 326 | 1.476 | 0.0388 | No | ||
| 2 | KPTN | 3520487 | 1568 | 0.448 | -0.0110 | No | ||
| 3 | LTB4R | 1770056 | 1775 | 0.384 | -0.0075 | No | ||
| 4 | ARPC1B | 5080736 5910136 | 1923 | 0.339 | -0.0025 | No | ||
| 5 | RAC1 | 4810687 | 1942 | 0.331 | 0.0093 | No | ||
| 6 | ARPC4 | 2900056 | 2004 | 0.317 | 0.0181 | No | ||
| 7 | CCL3 | 2810092 | 2095 | 0.290 | 0.0243 | No | ||
| 8 | MMP12 | 4920070 | 2263 | 0.252 | 0.0249 | No | ||
| 9 | CEECAM1 | 780605 | 2343 | 0.234 | 0.0296 | No | ||
| 10 | TNFRSF12A | 4810253 | 2508 | 0.201 | 0.0284 | No | ||
| 11 | IGF1 | 1990193 3130377 3290280 | 2798 | 0.153 | 0.0187 | No | ||
| 12 | AAMP | 2640059 | 2930 | 0.135 | 0.0168 | No | ||
| 13 | TPBG | 630161 | 2956 | 0.133 | 0.0205 | No | ||
| 14 | NEO1 | 1990154 | 2967 | 0.132 | 0.0250 | No | ||
| 15 | YARS | 70739 | 3021 | 0.126 | 0.0270 | No | ||
| 16 | NTF3 | 5890131 | 3097 | 0.118 | 0.0274 | No | ||
| 17 | MAPK8 | 2640195 | 3135 | 0.114 | 0.0298 | No | ||
| 18 | WASF1 | 2060484 | 3888 | 0.059 | -0.0086 | No | ||
| 19 | JAK2 | 3780528 6100692 6550577 | 4243 | 0.044 | -0.0260 | No | ||
| 20 | NOS3 | 630152 670465 | 4451 | 0.038 | -0.0357 | No | ||
| 21 | IL8RB | 450592 1170537 | 4641 | 0.033 | -0.0447 | No | ||
| 22 | THBS3 | 2360465 4050176 | 4710 | 0.031 | -0.0472 | No | ||
| 23 | CTGF | 4540577 | 4717 | 0.031 | -0.0463 | No | ||
| 24 | PYY | 2260546 | 4779 | 0.029 | -0.0485 | No | ||
| 25 | HRAS | 1980551 | 4798 | 0.029 | -0.0483 | No | ||
| 26 | XLKD1 | 520441 | 4843 | 0.028 | -0.0496 | No | ||
| 27 | HMMR | 5720315 6380168 | 4989 | 0.026 | -0.0564 | No | ||
| 28 | IL13 | 7040021 | 5371 | 0.021 | -0.0762 | No | ||
| 29 | NTN1 | 5700600 | 5549 | 0.019 | -0.0851 | No | ||
| 30 | FN1 | 1170601 2970647 6220288 6940037 | 5872 | 0.016 | -0.1019 | No | ||
| 31 | F11R | 1740128 | 6165 | 0.014 | -0.1171 | No | ||
| 32 | PTGS2 | 2510301 3170369 | 6247 | 0.013 | -0.1210 | No | ||
| 33 | SERPINB5 | 6940050 | 6351 | 0.012 | -0.1261 | No | ||
| 34 | HGFAC | 2510148 | 6526 | 0.011 | -0.1350 | No | ||
| 35 | OR1D2 | 3130333 | 6835 | 0.010 | -0.1513 | No | ||
| 36 | CHST4 | 1190008 | 7077 | 0.009 | -0.1640 | No | ||
| 37 | SERPINI2 | 1570427 | 7611 | 0.007 | -0.1925 | No | ||
| 38 | THBS4 | 3710348 | 7846 | 0.006 | -0.2049 | No | ||
| 39 | ANXA1 | 2320053 | 7855 | 0.006 | -0.2051 | No | ||
| 40 | AKAP4 | 2190731 3520619 | 7920 | 0.006 | -0.2084 | No | ||
| 41 | UBE3A | 1240152 2690438 5860609 | 7987 | 0.005 | -0.2117 | No | ||
| 42 | MAP2K1 | 840739 | 8060 | 0.005 | -0.2154 | No | ||
| 43 | DNALI1 | 3610402 | 8250 | 0.004 | -0.2255 | No | ||
| 44 | ADRA2A | 5340520 | 8620 | 0.003 | -0.2453 | No | ||
| 45 | DNAH5 | 7040315 | 8781 | 0.003 | -0.2538 | No | ||
| 46 | PAFAH1B2 | 5360546 6860672 | 8816 | 0.003 | -0.2555 | No | ||
| 47 | C3AR1 | 5720131 | 9078 | 0.002 | -0.2695 | No | ||
| 48 | CSPG4 | 2970270 1190148 | 9292 | 0.001 | -0.2810 | No | ||
| 49 | CXCL10 | 2450408 | 9427 | 0.001 | -0.2882 | No | ||
| 50 | HGF | 3360593 | 9670 | 0.000 | -0.3012 | No | ||
| 51 | VNN1 | 3940400 | 9823 | -0.000 | -0.3094 | No | ||
| 52 | THBS1 | 4560494 430288 | 10006 | -0.001 | -0.3193 | No | ||
| 53 | ENPP2 | 5860546 | 10195 | -0.001 | -0.3294 | No | ||
| 54 | ACTR3 | 1400497 | 10238 | -0.001 | -0.3316 | No | ||
| 55 | FPR1 | 3800577 | 10332 | -0.002 | -0.3365 | No | ||
| 56 | AKAP3 | 5890471 | 10445 | -0.002 | -0.3425 | No | ||
| 57 | MKLN1 | 70504 1850129 | 10536 | -0.002 | -0.3473 | No | ||
| 58 | FPRL1 | 3360451 | 11425 | -0.005 | -0.3951 | No | ||
| 59 | NPY | 3170138 | 11694 | -0.006 | -0.4093 | No | ||
| 60 | IFNG | 5670592 | 11695 | -0.006 | -0.4091 | No | ||
| 61 | CAPZA1 | 3140390 | 12655 | -0.010 | -0.4605 | No | ||
| 62 | ACTR2 | 1170332 | 13137 | -0.013 | -0.4859 | No | ||
| 63 | ARPC5 | 1740411 3450300 | 13625 | -0.018 | -0.5116 | No | ||
| 64 | CAPZA2 | 110427 4590064 | 13855 | -0.021 | -0.5231 | No | ||
| 65 | PAFAH1B1 | 4230333 6420121 6450066 | 14002 | -0.023 | -0.5301 | No | ||
| 66 | SEMA3E | 4230563 | 14006 | -0.023 | -0.5294 | No | ||
| 67 | CCL4 | 50368 430047 | 14237 | -0.027 | -0.5408 | No | ||
| 68 | GNA13 | 4590102 | 14479 | -0.032 | -0.5526 | No | ||
| 69 | BLR1 | 1500300 5420017 | 14670 | -0.037 | -0.5615 | No | ||
| 70 | ARFIP2 | 1850301 4670180 | 15091 | -0.058 | -0.5819 | No | ||
| 71 | PALM | 130601 3850017 | 15155 | -0.061 | -0.5830 | No | ||
| 72 | SPINT2 | 2630039 6110519 | 15408 | -0.081 | -0.5935 | No | ||
| 73 | LIMK1 | 5080064 | 15739 | -0.114 | -0.6070 | No | ||
| 74 | VASP | 7050500 | 16314 | -0.188 | -0.6308 | Yes | ||
| 75 | MST1R | 4050113 4610025 | 16359 | -0.196 | -0.6256 | Yes | ||
| 76 | CRK | 1230162 4780128 | 16406 | -0.206 | -0.6203 | Yes | ||
| 77 | PECAM1 | 4810139 | 16445 | -0.213 | -0.6142 | Yes | ||
| 78 | MAPK14 | 5290731 | 16518 | -0.228 | -0.6093 | Yes | ||
| 79 | ARPC2 | 6020484 | 16707 | -0.272 | -0.6091 | Yes | ||
| 80 | PAK4 | 1690152 | 16865 | -0.310 | -0.6057 | Yes | ||
| 81 | WASL | 3290170 | 17129 | -0.394 | -0.6049 | Yes | ||
| 82 | CXCR3 | 6590114 | 17204 | -0.414 | -0.5930 | Yes | ||
| 83 | BCL9L | 6940053 | 17235 | -0.427 | -0.5783 | Yes | ||
| 84 | MARCKS | 7040450 | 17258 | -0.436 | -0.5628 | Yes | ||
| 85 | ACTN4 | 3840301 4590390 7050132 | 17404 | -0.498 | -0.5516 | Yes | ||
| 86 | ARPC3 | 670441 | 17421 | -0.505 | -0.5331 | Yes | ||
| 87 | AMFR | 2810041 | 17695 | -0.655 | -0.5228 | Yes | ||
| 88 | FLNA | 5390193 | 17743 | -0.680 | -0.4994 | Yes | ||
| 89 | MSN | 4060044 | 17745 | -0.681 | -0.4734 | Yes | ||
| 90 | CRKL | 4050427 | 17897 | -0.783 | -0.4516 | Yes | ||
| 91 | ITGAL | 1450324 | 17929 | -0.809 | -0.4223 | Yes | ||
| 92 | STAT3 | 460040 3710341 | 18021 | -0.893 | -0.3931 | Yes | ||
| 93 | CD97 | 1090673 1690575 | 18123 | -1.017 | -0.3597 | Yes | ||
| 94 | SELL | 1190500 | 18127 | -1.022 | -0.3208 | Yes | ||
| 95 | F2R | 4810180 | 18214 | -1.148 | -0.2815 | Yes | ||
| 96 | CCL5 | 3710397 | 18504 | -2.172 | -0.2140 | Yes | ||
| 97 | CORO1A | 3140609 3190020 3190037 | 18534 | -2.484 | -0.1206 | Yes | ||
| 98 | LSP1 | 2350091 | 18586 | -3.268 | 0.0016 | Yes |

