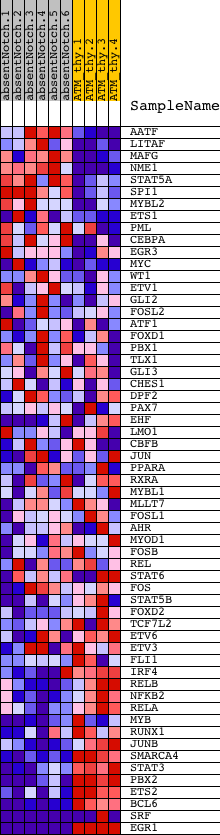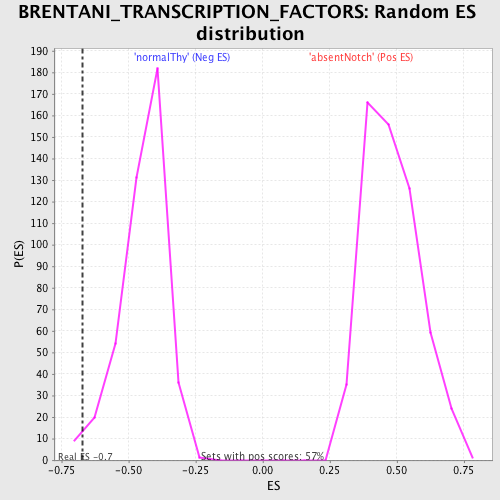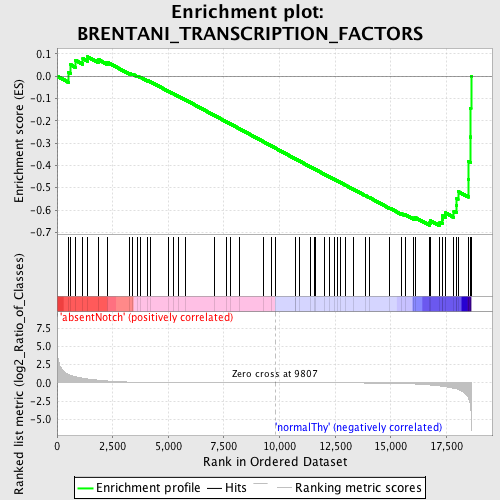
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | BRENTANI_TRANSCRIPTION_FACTORS |
| Enrichment Score (ES) | -0.67151666 |
| Normalized Enrichment Score (NES) | -1.5115081 |
| Nominal p-value | 0.016166281 |
| FDR q-value | 0.44303825 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AATF | 5220575 | 518 | 1.134 | 0.0158 | No | ||
| 2 | LITAF | 6940671 | 598 | 1.042 | 0.0517 | No | ||
| 3 | MAFG | 4120440 | 814 | 0.833 | 0.0722 | No | ||
| 4 | NME1 | 770014 | 1159 | 0.629 | 0.0780 | No | ||
| 5 | STAT5A | 2680458 | 1375 | 0.523 | 0.0865 | No | ||
| 6 | SPI1 | 1410397 | 1873 | 0.353 | 0.0734 | No | ||
| 7 | MYBL2 | 5220609 6520368 | 2253 | 0.253 | 0.0627 | No | ||
| 8 | ETS1 | 5270278 6450717 6620465 | 3268 | 0.100 | 0.0119 | No | ||
| 9 | PML | 50093 2190435 2450402 3840082 | 3377 | 0.092 | 0.0097 | No | ||
| 10 | CEBPA | 5690519 | 3602 | 0.076 | 0.0005 | No | ||
| 11 | EGR3 | 6940128 | 3745 | 0.067 | -0.0045 | No | ||
| 12 | MYC | 380541 4670170 | 4075 | 0.051 | -0.0203 | No | ||
| 13 | WT1 | 3990463 | 4204 | 0.046 | -0.0254 | No | ||
| 14 | ETV1 | 70735 2940603 5080463 | 5007 | 0.026 | -0.0677 | No | ||
| 15 | GLI2 | 3060632 | 5212 | 0.022 | -0.0778 | No | ||
| 16 | FOSL2 | 870044 1230427 6520433 | 5454 | 0.020 | -0.0900 | No | ||
| 17 | ATF1 | 4570142 5550095 | 5748 | 0.017 | -0.1052 | No | ||
| 18 | FOXD1 | 6860053 | 7059 | 0.009 | -0.1754 | No | ||
| 19 | PBX1 | 6660301 | 7635 | 0.007 | -0.2062 | No | ||
| 20 | TLX1 | 2630162 | 7776 | 0.006 | -0.2135 | No | ||
| 21 | GLI3 | 5690148 | 8188 | 0.005 | -0.2354 | No | ||
| 22 | CHES1 | 4570121 5700162 | 9295 | 0.001 | -0.2950 | No | ||
| 23 | DPF2 | 6980373 | 9631 | 0.001 | -0.3130 | No | ||
| 24 | PAX7 | 6860593 | 9812 | -0.000 | -0.3227 | No | ||
| 25 | EHF | 4560088 | 10719 | -0.003 | -0.3714 | No | ||
| 26 | LMO1 | 3170528 | 10730 | -0.003 | -0.3719 | No | ||
| 27 | CBFB | 1850551 2650035 | 10875 | -0.003 | -0.3795 | No | ||
| 28 | JUN | 840170 | 11404 | -0.005 | -0.4078 | No | ||
| 29 | PPARA | 2060026 | 11546 | -0.005 | -0.4152 | No | ||
| 30 | RXRA | 3800471 | 11617 | -0.006 | -0.4187 | No | ||
| 31 | MYBL1 | 2900168 | 12007 | -0.007 | -0.4394 | No | ||
| 32 | MLLT7 | 4480707 | 12258 | -0.008 | -0.4526 | No | ||
| 33 | FOSL1 | 430021 | 12472 | -0.009 | -0.4637 | No | ||
| 34 | AHR | 4560671 | 12618 | -0.010 | -0.4711 | No | ||
| 35 | MYOD1 | 1230731 | 12744 | -0.011 | -0.4774 | No | ||
| 36 | FOSB | 1940142 | 12947 | -0.012 | -0.4878 | No | ||
| 37 | REL | 360707 | 13310 | -0.015 | -0.5068 | No | ||
| 38 | STAT6 | 1190010 5720019 | 13858 | -0.021 | -0.5355 | No | ||
| 39 | FOS | 1850315 | 14022 | -0.023 | -0.5433 | No | ||
| 40 | STAT5B | 6200026 | 14943 | -0.049 | -0.5910 | No | ||
| 41 | FOXD2 | 4210035 | 15489 | -0.088 | -0.6170 | No | ||
| 42 | TCF7L2 | 2190348 5220113 | 15650 | -0.105 | -0.6216 | No | ||
| 43 | ETV6 | 610524 | 16017 | -0.145 | -0.6357 | No | ||
| 44 | ETV3 | 5360487 | 16114 | -0.160 | -0.6347 | No | ||
| 45 | FLI1 | 3990142 | 16745 | -0.280 | -0.6579 | Yes | ||
| 46 | IRF4 | 610133 | 16775 | -0.287 | -0.6484 | Yes | ||
| 47 | RELB | 1400048 | 17205 | -0.414 | -0.6555 | Yes | ||
| 48 | NFKB2 | 2320670 | 17315 | -0.460 | -0.6437 | Yes | ||
| 49 | RELA | 3830075 | 17328 | -0.467 | -0.6263 | Yes | ||
| 50 | MYB | 1660494 5860451 6130706 | 17450 | -0.518 | -0.6129 | Yes | ||
| 51 | RUNX1 | 3840711 | 17835 | -0.736 | -0.6052 | Yes | ||
| 52 | JUNB | 4230048 | 17941 | -0.818 | -0.5793 | Yes | ||
| 53 | SMARCA4 | 6980528 | 17966 | -0.837 | -0.5484 | Yes | ||
| 54 | STAT3 | 460040 3710341 | 18021 | -0.893 | -0.5169 | Yes | ||
| 55 | PBX2 | 3130433 6510292 | 18485 | -2.018 | -0.4640 | Yes | ||
| 56 | ETS2 | 360451 | 18501 | -2.149 | -0.3820 | Yes | ||
| 57 | BCL6 | 940100 | 18572 | -2.937 | -0.2725 | Yes | ||
| 58 | SRF | 70139 2320039 | 18588 | -3.326 | -0.1451 | Yes | ||
| 59 | EGR1 | 4610347 | 18604 | -3.801 | 0.0006 | Yes |