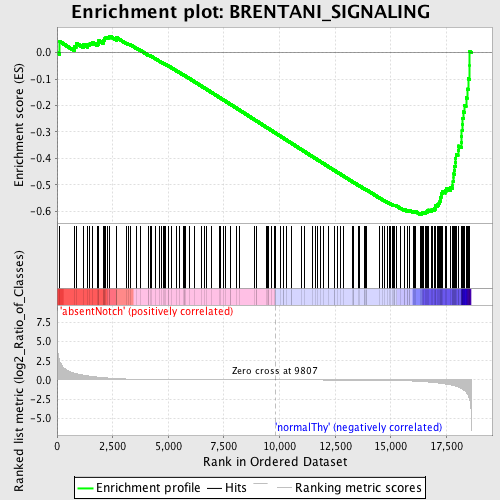
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | BRENTANI_SIGNALING |
| Enrichment Score (ES) | -0.6116679 |
| Normalized Enrichment Score (NES) | -1.5589404 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.35061792 |
| FWER p-Value | 0.977 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MAP3K7IP1 | 2120095 5890452 6040167 | 121 | 2.379 | 0.0419 | No | ||
| 2 | TNFSF13 | 2650170 | 784 | 0.851 | 0.0234 | No | ||
| 3 | MAPK13 | 6760215 | 853 | 0.811 | 0.0362 | No | ||
| 4 | AXL | 1770095 | 1174 | 0.619 | 0.0315 | No | ||
| 5 | STAT5A | 2680458 | 1375 | 0.523 | 0.0313 | No | ||
| 6 | GRB7 | 2100471 | 1477 | 0.484 | 0.0357 | No | ||
| 7 | IL12RB1 | 2230338 5420053 | 1582 | 0.443 | 0.0391 | No | ||
| 8 | MAP2K4 | 5130133 | 1821 | 0.367 | 0.0337 | No | ||
| 9 | MAPK10 | 6110193 | 1857 | 0.358 | 0.0391 | No | ||
| 10 | ITGB1 | 5080156 6270528 | 1874 | 0.353 | 0.0454 | No | ||
| 11 | SMO | 5340538 6350132 | 2069 | 0.301 | 0.0410 | No | ||
| 12 | ITGA9 | 670180 | 2096 | 0.290 | 0.0455 | No | ||
| 13 | MAPKAPK2 | 2850500 | 2120 | 0.284 | 0.0501 | No | ||
| 14 | CXCL1 | 2690537 | 2144 | 0.280 | 0.0545 | No | ||
| 15 | ITGB5 | 70129 | 2191 | 0.267 | 0.0575 | No | ||
| 16 | IL10 | 2340685 2640541 2850403 6590286 | 2268 | 0.251 | 0.0585 | No | ||
| 17 | JAG2 | 1500341 | 2335 | 0.236 | 0.0597 | No | ||
| 18 | RAF1 | 1770600 | 2375 | 0.227 | 0.0622 | No | ||
| 19 | TYRO3 | 3130193 | 2664 | 0.175 | 0.0502 | No | ||
| 20 | BTK | 3130044 | 2668 | 0.174 | 0.0536 | No | ||
| 21 | MAP2K2 | 4590601 | 2672 | 0.174 | 0.0570 | No | ||
| 22 | ABL1 | 1050593 2030050 4010114 | 3116 | 0.115 | 0.0353 | No | ||
| 23 | YWHAE | 5310435 | 3213 | 0.106 | 0.0322 | No | ||
| 24 | CAMK4 | 1690091 | 3297 | 0.098 | 0.0297 | No | ||
| 25 | NRAS | 6900577 | 3584 | 0.077 | 0.0158 | No | ||
| 26 | NOTCH4 | 2450040 6370707 | 3758 | 0.066 | 0.0078 | No | ||
| 27 | TNFSF4 | 3940594 | 4091 | 0.050 | -0.0092 | No | ||
| 28 | IL1B | 2640364 | 4117 | 0.049 | -0.0096 | No | ||
| 29 | MAPK1 | 3190193 6200253 | 4185 | 0.046 | -0.0122 | No | ||
| 30 | TANK | 1090020 2060458 2370400 | 4226 | 0.045 | -0.0135 | No | ||
| 31 | BCR | 2260020 4230180 6040195 | 4419 | 0.039 | -0.0231 | No | ||
| 32 | TNFRSF10B | 940671 1500451 6550768 6900500 | 4600 | 0.034 | -0.0322 | No | ||
| 33 | ITGA3 | 4570427 | 4687 | 0.032 | -0.0362 | No | ||
| 34 | HRAS | 1980551 | 4798 | 0.029 | -0.0416 | No | ||
| 35 | PTCH2 | 2900092 | 4818 | 0.029 | -0.0420 | No | ||
| 36 | TNF | 6650603 | 4887 | 0.028 | -0.0451 | No | ||
| 37 | ERG | 50154 1770739 | 5000 | 0.026 | -0.0507 | No | ||
| 38 | MCC | 620750 | 5028 | 0.025 | -0.0516 | No | ||
| 39 | CXCL2 | 610398 | 5151 | 0.023 | -0.0578 | No | ||
| 40 | IL13 | 7040021 | 5371 | 0.021 | -0.0692 | No | ||
| 41 | IFNB1 | 1400142 | 5487 | 0.019 | -0.0750 | No | ||
| 42 | ARHGEF16 | 3990372 | 5658 | 0.018 | -0.0839 | No | ||
| 43 | TNFRSF1B | 3990035 5860372 | 5722 | 0.017 | -0.0870 | No | ||
| 44 | CALM1 | 380128 | 5743 | 0.017 | -0.0877 | No | ||
| 45 | ITGA4 | 2470739 | 5778 | 0.016 | -0.0892 | No | ||
| 46 | MAP2K7 | 2260086 | 5971 | 0.015 | -0.0993 | No | ||
| 47 | INHA | 6100102 | 6190 | 0.013 | -0.1109 | No | ||
| 48 | WNT4 | 4150619 | 6493 | 0.012 | -0.1270 | No | ||
| 49 | SUFU | 6650181 | 6619 | 0.011 | -0.1336 | No | ||
| 50 | TTK | 3800129 | 6736 | 0.010 | -0.1396 | No | ||
| 51 | ARHGAP6 | 2060121 | 6945 | 0.009 | -0.1507 | No | ||
| 52 | MPL | 2360253 | 7295 | 0.008 | -0.1695 | No | ||
| 53 | MAP3K4 | 5720041 | 7343 | 0.008 | -0.1719 | No | ||
| 54 | IGFBP3 | 2370500 | 7467 | 0.007 | -0.1784 | No | ||
| 55 | MAPK11 | 130452 3120440 3610465 | 7589 | 0.007 | -0.1848 | No | ||
| 56 | AKT1 | 5290746 | 7797 | 0.006 | -0.1959 | No | ||
| 57 | ITGA7 | 1230739 | 7800 | 0.006 | -0.1959 | No | ||
| 58 | MAP2K1 | 840739 | 8060 | 0.005 | -0.2098 | No | ||
| 59 | RAP1A | 1090025 | 8182 | 0.005 | -0.2163 | No | ||
| 60 | ARHGEF12 | 3990195 | 8868 | 0.003 | -0.2534 | No | ||
| 61 | SKIL | 1090364 | 8954 | 0.002 | -0.2579 | No | ||
| 62 | WNT1 | 4780148 | 8974 | 0.002 | -0.2589 | No | ||
| 63 | ROS1 | 1090019 | 9403 | 0.001 | -0.2821 | No | ||
| 64 | ITGAM | 1190373 | 9446 | 0.001 | -0.2843 | No | ||
| 65 | MAS1 | 2650600 | 9500 | 0.001 | -0.2872 | No | ||
| 66 | GRB14 | 510035 3060369 | 9508 | 0.001 | -0.2876 | No | ||
| 67 | ITGAV | 2340020 | 9652 | 0.000 | -0.2953 | No | ||
| 68 | RAN | 2260446 4590647 | 9782 | 0.000 | -0.3023 | No | ||
| 69 | MAPKAPK5 | 2510601 | 9793 | 0.000 | -0.3028 | No | ||
| 70 | MAP3K12 | 2470373 6420162 | 9831 | -0.000 | -0.3048 | No | ||
| 71 | WNT5A | 840685 3120152 | 10055 | -0.001 | -0.3169 | No | ||
| 72 | IGFBP1 | 5720435 | 10169 | -0.001 | -0.3230 | No | ||
| 73 | MUSK | 1190113 | 10173 | -0.001 | -0.3231 | No | ||
| 74 | MAPK7 | 5670079 6100128 | 10297 | -0.001 | -0.3298 | No | ||
| 75 | MAPK12 | 450022 1340717 7050484 | 10517 | -0.002 | -0.3416 | No | ||
| 76 | ABL2 | 580021 | 10531 | -0.002 | -0.3423 | No | ||
| 77 | ITGAX | 6130524 | 11003 | -0.004 | -0.3677 | No | ||
| 78 | SOS1 | 7050338 | 11108 | -0.004 | -0.3733 | No | ||
| 79 | MAPK4 | 870142 | 11473 | -0.005 | -0.3929 | No | ||
| 80 | WNT2 | 4610020 | 11616 | -0.006 | -0.4005 | No | ||
| 81 | IFNG | 5670592 | 11695 | -0.006 | -0.4046 | No | ||
| 82 | APC | 3850484 5860722 | 11816 | -0.006 | -0.4110 | No | ||
| 83 | ATM | 3610110 4050524 | 11982 | -0.007 | -0.4198 | No | ||
| 84 | JAG1 | 3440390 | 12186 | -0.008 | -0.4306 | No | ||
| 85 | NCK1 | 6200575 6510050 | 12455 | -0.009 | -0.4450 | No | ||
| 86 | SRC | 580132 | 12478 | -0.009 | -0.4460 | No | ||
| 87 | AKT2 | 3850541 3870519 | 12620 | -0.010 | -0.4534 | No | ||
| 88 | NF1 | 6980433 | 12719 | -0.011 | -0.4585 | No | ||
| 89 | SYK | 6940133 | 12885 | -0.012 | -0.4672 | No | ||
| 90 | IRS1 | 1190204 | 13277 | -0.014 | -0.4881 | No | ||
| 91 | REL | 360707 | 13310 | -0.015 | -0.4895 | No | ||
| 92 | MAP3K5 | 6020041 6380162 | 13343 | -0.015 | -0.4910 | No | ||
| 93 | IL15 | 4560332 5860438 | 13562 | -0.017 | -0.5024 | No | ||
| 94 | BIRC2 | 2940435 4730020 | 13581 | -0.017 | -0.5031 | No | ||
| 95 | PTPRK | 1450433 2640142 | 13831 | -0.020 | -0.5161 | No | ||
| 96 | IFNAR1 | 4730672 | 13832 | -0.020 | -0.5157 | No | ||
| 97 | STAT6 | 1190010 5720019 | 13858 | -0.021 | -0.5167 | No | ||
| 98 | SHC1 | 2900731 3170504 6520537 | 13907 | -0.021 | -0.5188 | No | ||
| 99 | DLG1 | 2630091 6020286 | 13917 | -0.022 | -0.5189 | No | ||
| 100 | VIPR1 | 2480292 5720288 6760537 | 14482 | -0.032 | -0.5488 | No | ||
| 101 | ARHGAP1 | 2810010 5270064 | 14636 | -0.036 | -0.5563 | No | ||
| 102 | ITGB4 | 1740021 3840482 | 14703 | -0.038 | -0.5591 | No | ||
| 103 | CXCL12 | 580546 4150750 4570068 | 14869 | -0.045 | -0.5672 | No | ||
| 104 | STAT5B | 6200026 | 14943 | -0.049 | -0.5701 | No | ||
| 105 | ITGB7 | 4670619 | 14968 | -0.050 | -0.5704 | No | ||
| 106 | ILK | 870601 4480180 | 15077 | -0.057 | -0.5751 | No | ||
| 107 | SFN | 6290301 7510608 | 15114 | -0.059 | -0.5759 | No | ||
| 108 | JAK3 | 70347 3290008 | 15166 | -0.062 | -0.5774 | No | ||
| 109 | ITGA5 | 5550520 | 15174 | -0.063 | -0.5765 | No | ||
| 110 | ITGB3 | 5270463 | 15243 | -0.068 | -0.5788 | No | ||
| 111 | IL18R1 | 270546 | 15434 | -0.083 | -0.5874 | No | ||
| 112 | IKBKG | 3450092 3840377 6590592 | 15620 | -0.102 | -0.5953 | No | ||
| 113 | ITGAE | 4210632 4670239 5340253 5420746 5690154 5700685 6770154 | 15630 | -0.103 | -0.5937 | No | ||
| 114 | TIAM1 | 5420288 | 15730 | -0.113 | -0.5968 | No | ||
| 115 | FYN | 2100468 4760520 4850687 | 15761 | -0.116 | -0.5960 | No | ||
| 116 | MAP3K8 | 2940286 | 15842 | -0.125 | -0.5978 | No | ||
| 117 | JAK1 | 5910746 | 15852 | -0.127 | -0.5957 | No | ||
| 118 | ROCK1 | 130044 | 16034 | -0.147 | -0.6025 | No | ||
| 119 | ZAP70 | 1410494 2260504 | 16061 | -0.152 | -0.6008 | No | ||
| 120 | MAP2K3 | 5570193 | 16103 | -0.158 | -0.5998 | No | ||
| 121 | MAP4K4 | 2360059 | 16313 | -0.188 | -0.6073 | No | ||
| 122 | SH3BP2 | 610131 | 16394 | -0.203 | -0.6075 | Yes | ||
| 123 | CRK | 1230162 4780128 | 16406 | -0.206 | -0.6039 | Yes | ||
| 124 | TYK2 | 1660465 1740736 | 16473 | -0.219 | -0.6030 | Yes | ||
| 125 | MAP2K5 | 2030440 | 16563 | -0.240 | -0.6030 | Yes | ||
| 126 | TRAF6 | 4810292 6200132 | 16620 | -0.253 | -0.6009 | Yes | ||
| 127 | MKNK1 | 6510161 | 16654 | -0.259 | -0.5974 | Yes | ||
| 128 | RGS16 | 780091 | 16708 | -0.272 | -0.5947 | Yes | ||
| 129 | MYD88 | 2100717 | 16819 | -0.299 | -0.5945 | Yes | ||
| 130 | IL2RB | 4730072 | 16889 | -0.317 | -0.5918 | Yes | ||
| 131 | RASA1 | 1240315 | 16954 | -0.335 | -0.5885 | Yes | ||
| 132 | PIK3C3 | 6590717 | 17008 | -0.350 | -0.5842 | Yes | ||
| 133 | IL6ST | 5890047 | 17016 | -0.352 | -0.5774 | Yes | ||
| 134 | GNB1 | 2120397 | 17111 | -0.389 | -0.5746 | Yes | ||
| 135 | PTEN | 3390064 | 17161 | -0.403 | -0.5691 | Yes | ||
| 136 | RELB | 1400048 | 17205 | -0.414 | -0.5629 | Yes | ||
| 137 | AXIN2 | 3850131 | 17220 | -0.423 | -0.5551 | Yes | ||
| 138 | TNFRSF4 | 6840056 | 17224 | -0.423 | -0.5466 | Yes | ||
| 139 | MAP2K6 | 1230056 2940204 | 17261 | -0.436 | -0.5397 | Yes | ||
| 140 | RALBP1 | 4780632 | 17268 | -0.439 | -0.5311 | Yes | ||
| 141 | RELA | 3830075 | 17328 | -0.467 | -0.5247 | Yes | ||
| 142 | ITGA6 | 3830129 | 17438 | -0.512 | -0.5202 | Yes | ||
| 143 | MAP3K14 | 5890435 | 17492 | -0.536 | -0.5122 | Yes | ||
| 144 | STAT2 | 3780519 | 17679 | -0.645 | -0.5091 | Yes | ||
| 145 | BCL10 | 2360397 | 17788 | -0.705 | -0.5006 | Yes | ||
| 146 | AXIN1 | 3360358 6940451 | 17793 | -0.709 | -0.4864 | Yes | ||
| 147 | ITGB2 | 6380348 | 17832 | -0.734 | -0.4735 | Yes | ||
| 148 | PIK3CG | 5890110 | 17834 | -0.735 | -0.4585 | Yes | ||
| 149 | VAV1 | 6020487 | 17855 | -0.751 | -0.4443 | Yes | ||
| 150 | TNFRSF14 | 5050438 | 17871 | -0.760 | -0.4296 | Yes | ||
| 151 | CRKL | 4050427 | 17897 | -0.783 | -0.4150 | Yes | ||
| 152 | DGKQ | 2480609 | 17918 | -0.799 | -0.3998 | Yes | ||
| 153 | ITGAL | 1450324 | 17929 | -0.809 | -0.3839 | Yes | ||
| 154 | STAT3 | 460040 3710341 | 18021 | -0.893 | -0.3706 | Yes | ||
| 155 | TRAF1 | 3440735 | 18036 | -0.911 | -0.3528 | Yes | ||
| 156 | STAT1 | 6510204 6590553 | 18176 | -1.089 | -0.3382 | Yes | ||
| 157 | MAP3K3 | 610685 | 18187 | -1.104 | -0.3162 | Yes | ||
| 158 | TRAF3 | 5690647 | 18198 | -1.124 | -0.2938 | Yes | ||
| 159 | NOTCH3 | 3060603 | 18226 | -1.164 | -0.2716 | Yes | ||
| 160 | IFNGR1 | 610577 | 18234 | -1.174 | -0.2480 | Yes | ||
| 161 | PDPK1 | 6650168 | 18263 | -1.250 | -0.2241 | Yes | ||
| 162 | IFNAR2 | 4070687 7100484 | 18316 | -1.364 | -0.1991 | Yes | ||
| 163 | PTK2B | 4730411 | 18382 | -1.574 | -0.1705 | Yes | ||
| 164 | MAPK9 | 2060273 3780209 4070397 | 18430 | -1.744 | -0.1376 | Yes | ||
| 165 | DGKA | 5720152 5890328 | 18492 | -2.053 | -0.0990 | Yes | ||
| 166 | DGKG | 1770025 | 18546 | -2.567 | -0.0496 | Yes | ||
| 167 | TNFRSF1A | 1090390 6520735 | 18550 | -2.617 | 0.0036 | Yes |

