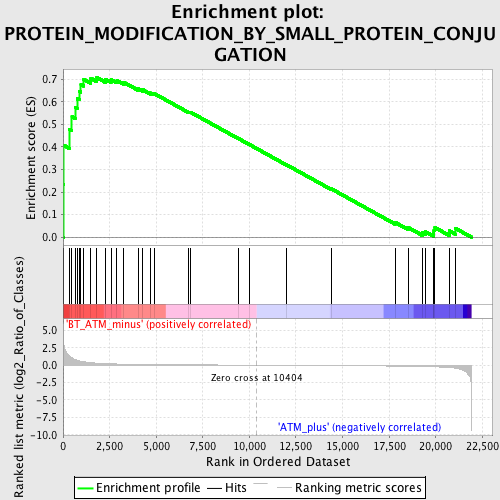
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | PROTEIN_MODIFICATION_BY_SMALL_PROTEIN_CONJUGATION |
| Enrichment Score (ES) | 0.7071346 |
| Normalized Enrichment Score (NES) | 1.642835 |
| Nominal p-value | 0.010948905 |
| FDR q-value | 0.6568943 |
| FWER p-Value | 0.995 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UHRF2 | 1447217_at 1454920_at 1457070_at | 17 | 3.839 | 0.2343 | Yes | ||
| 2 | RNF139 | 1429425_at 1429426_at | 45 | 2.841 | 0.4070 | Yes | ||
| 3 | CBL | 1434829_at 1446608_at 1450457_at 1455886_at | 334 | 1.363 | 0.4773 | Yes | ||
| 4 | ANAPC11 | 1417326_a_at 1447897_x_at 1459212_at | 476 | 1.046 | 0.5349 | Yes | ||
| 5 | SENP7 | 1430974_a_at 1436860_at 1438413_at | 647 | 0.801 | 0.5762 | Yes | ||
| 6 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 754 | 0.697 | 0.6140 | Yes | ||
| 7 | PCNP | 1428314_at 1452735_at | 867 | 0.611 | 0.6463 | Yes | ||
| 8 | UBE2V2 | 1417983_a_at 1417984_at 1429131_at 1429132_at | 958 | 0.562 | 0.6766 | Yes | ||
| 9 | STUB1 | 1416580_a_at 1429069_at 1434412_x_at | 1111 | 0.487 | 0.6994 | Yes | ||
| 10 | UBE2N | 1422559_at 1435384_at | 1491 | 0.354 | 0.7038 | Yes | ||
| 11 | PARK2 | 1420755_a_at 1426135_a_at 1449975_a_at | 1811 | 0.292 | 0.7071 | Yes | ||
| 12 | GTPBP4 | 1423142_a_at 1423143_at 1450873_at | 2269 | 0.226 | 0.7001 | No | ||
| 13 | WWP1 | 1427097_at 1427098_at 1446364_at 1446853_at 1452299_at | 2576 | 0.193 | 0.6980 | No | ||
| 14 | FBXW7 | 1424986_s_at 1451558_at | 2892 | 0.168 | 0.6939 | No | ||
| 15 | SMURF1 | 1443393_at | 3256 | 0.147 | 0.6863 | No | ||
| 16 | UBE2C | 1452954_at | 4036 | 0.115 | 0.6577 | No | ||
| 17 | ATG3 | 1418179_at 1448993_at | 4285 | 0.106 | 0.6529 | No | ||
| 18 | TRIM23 | 1426969_at 1429998_at 1456089_at | 4714 | 0.094 | 0.6391 | No | ||
| 19 | TSC1 | 1422043_at 1439989_at 1455252_at | 4910 | 0.089 | 0.6357 | No | ||
| 20 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 6756 | 0.052 | 0.5546 | No | ||
| 21 | HUWE1 | 1415703_at 1424949_at 1443183_at 1443649_at 1444279_at 1448056_at | 6848 | 0.051 | 0.5536 | No | ||
| 22 | TRAF7 | 1424320_a_at | 9397 | 0.013 | 0.4381 | No | ||
| 23 | SUMO1 | 1438289_a_at 1444557_at 1451005_at 1456349_x_at | 10014 | 0.005 | 0.4103 | No | ||
| 24 | ERCC8 | 1427194_a_at 1429439_at 1432249_a_at 1454176_at | 12009 | -0.021 | 0.3205 | No | ||
| 25 | BRCA1 | 1424629_at 1424630_a_at 1427698_at 1443763_at 1451417_at | 14398 | -0.055 | 0.2148 | No | ||
| 26 | RNF11 | 1426404_a_at 1426405_at 1439685_at 1442048_at 1452058_a_at | 17828 | -0.128 | 0.0661 | No | ||
| 27 | MYLIP | 1424988_at | 18537 | -0.151 | 0.0430 | No | ||
| 28 | DDB2 | 1425706_a_at 1436680_s_at | 19294 | -0.185 | 0.0198 | No | ||
| 29 | UBE4B | 1432499_a_at 1444465_at 1448410_at | 19458 | -0.194 | 0.0242 | No | ||
| 30 | TRAF6 | 1421376_at 1421377_at 1435350_at 1443288_at 1446940_at 1446977_at | 19872 | -0.224 | 0.0191 | No | ||
| 31 | UBE2D1 | 1424062_at | 19913 | -0.229 | 0.0313 | No | ||
| 32 | AMFR | 1450102_a_at | 19966 | -0.232 | 0.0431 | No | ||
| 33 | UBE3C | 1426881_at 1426882_at 1444562_at 1448097_at 1456992_at 1458637_x_at | 20738 | -0.334 | 0.0284 | No | ||
| 34 | CAND1 | 1428648_at 1428649_at 1435781_at 1455751_at | 21062 | -0.428 | 0.0398 | No |

