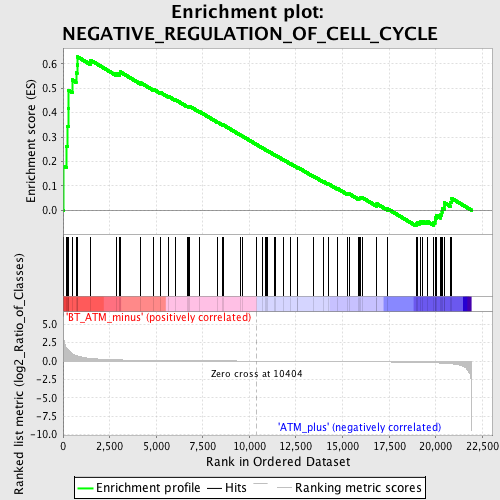
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | NEGATIVE_REGULATION_OF_CELL_CYCLE |
| Enrichment Score (ES) | 0.6286397 |
| Normalized Enrichment Score (NES) | 1.6176901 |
| Nominal p-value | 0.008888889 |
| FDR q-value | 0.48895243 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PPP1R13B | 1434895_s_at 1442956_at 1459224_at | 22 | 3.608 | 0.1813 | Yes | ||
| 2 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 192 | 1.767 | 0.2628 | Yes | ||
| 3 | RASSF1 | 1441737_s_at 1448855_at 1456994_at 1457788_at | 226 | 1.646 | 0.3445 | Yes | ||
| 4 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 276 | 1.510 | 0.4185 | Yes | ||
| 5 | TGFB1 | 1420653_at 1445360_at | 303 | 1.452 | 0.4906 | Yes | ||
| 6 | CUL3 | 1422794_at 1422795_at 1434717_at 1434718_at 1450756_s_at | 489 | 1.033 | 0.5343 | Yes | ||
| 7 | EIF4G2 | 1415863_at 1452758_s_at | 696 | 0.753 | 0.5629 | Yes | ||
| 8 | ING4 | 1448054_at 1460728_s_at | 750 | 0.700 | 0.5959 | Yes | ||
| 9 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 779 | 0.674 | 0.6286 | Yes | ||
| 10 | MAP2K6 | 1426850_a_at 1459292_at | 1462 | 0.361 | 0.6157 | No | ||
| 11 | CUL1 | 1448174_at 1459516_at | 2861 | 0.171 | 0.5604 | No | ||
| 12 | CDKN3 | 1415968_a_at 1415969_s_at 1430574_at | 3048 | 0.158 | 0.5599 | No | ||
| 13 | LATS1 | 1427679_at 1460264_at | 3061 | 0.158 | 0.5673 | No | ||
| 14 | MFN2 | 1441424_at 1448131_at 1451900_at | 4164 | 0.109 | 0.5224 | No | ||
| 15 | NBN | 1448746_at | 4836 | 0.091 | 0.4963 | No | ||
| 16 | BMP4 | 1422912_at | 5231 | 0.082 | 0.4825 | No | ||
| 17 | UHMK1 | 1446120_at 1454260_at | 5682 | 0.072 | 0.4655 | No | ||
| 18 | HEXIM1 | 1419359_at 1425937_a_at | 6043 | 0.065 | 0.4524 | No | ||
| 19 | PPP1R9B | 1447247_at 1447486_at | 6657 | 0.054 | 0.4271 | No | ||
| 20 | BMP7 | 1418910_at 1432410_a_at | 6752 | 0.052 | 0.4254 | No | ||
| 21 | CDKN2D | 1416253_at | 6802 | 0.052 | 0.4258 | No | ||
| 22 | CDKN1B | 1419497_at 1434045_at | 7343 | 0.042 | 0.4032 | No | ||
| 23 | FOXC1 | 1419485_at 1419486_at | 8315 | 0.028 | 0.3603 | No | ||
| 24 | BMP2 | 1423635_at | 8557 | 0.025 | 0.3505 | No | ||
| 25 | RPRM | 1422552_at | 8597 | 0.024 | 0.3499 | No | ||
| 26 | GAS1 | 1416855_at 1448494_at 1456322_at | 9519 | 0.012 | 0.3084 | No | ||
| 27 | GADD45A | 1449519_at | 9649 | 0.010 | 0.3030 | No | ||
| 28 | HPGD | 1419905_s_at 1419906_at | 10366 | 0.001 | 0.2703 | No | ||
| 29 | INHA | 1422728_at | 10713 | -0.004 | 0.2547 | No | ||
| 30 | PCAF | 1434037_s_at 1444473_at 1445105_at 1450821_at | 10848 | -0.006 | 0.2488 | No | ||
| 31 | STK11 | 1418644_a_at | 10920 | -0.007 | 0.2459 | No | ||
| 32 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 10982 | -0.008 | 0.2435 | No | ||
| 33 | CDKN2A | 1450140_a_at | 11368 | -0.013 | 0.2266 | No | ||
| 34 | ZBTB17 | 1416224_at 1443041_at | 11425 | -0.013 | 0.2247 | No | ||
| 35 | CUL4A | 1451971_at | 11858 | -0.019 | 0.2059 | No | ||
| 36 | HEXIM2 | 1429025_a_at 1429026_at | 12227 | -0.024 | 0.1902 | No | ||
| 37 | ERN1 | 1421240_at 1431886_at 1437119_at 1438583_at 1438997_at 1450176_at | 12605 | -0.029 | 0.1745 | No | ||
| 38 | CUL2 | 1419765_at 1451231_a_at | 12608 | -0.029 | 0.1758 | No | ||
| 39 | JMY | 1420639_at 1420640_at 1451878_a_at | 13445 | -0.041 | 0.1396 | No | ||
| 40 | CDKN2C | 1416868_at 1439164_at | 13994 | -0.049 | 0.1171 | No | ||
| 41 | MAPK12 | 1447823_x_at 1449283_a_at | 14253 | -0.053 | 0.1079 | No | ||
| 42 | PPM1G | 1416792_at | 14731 | -0.061 | 0.0892 | No | ||
| 43 | APBB1 | 1423892_at 1423893_x_at | 15257 | -0.069 | 0.0687 | No | ||
| 44 | LATS2 | 1419678_at 1419679_at 1425420_s_at 1426156_at 1437101_at 1439441_x_at | 15367 | -0.071 | 0.0673 | No | ||
| 45 | CUL5 | 1428287_at 1452722_a_at 1456102_a_at | 15852 | -0.080 | 0.0492 | No | ||
| 46 | GAS7 | 1417859_at 1431400_a_at 1445685_at | 15917 | -0.081 | 0.0503 | No | ||
| 47 | PLAGL1 | 1426208_x_at 1438588_at 1450533_a_at | 15943 | -0.082 | 0.0533 | No | ||
| 48 | CDKN2B | 1449152_at | 16050 | -0.083 | 0.0527 | No | ||
| 49 | RHOB | 1449110_at | 16837 | -0.101 | 0.0219 | No | ||
| 50 | MLF1 | 1418589_a_at | 16852 | -0.101 | 0.0263 | No | ||
| 51 | BTG4 | 1426520_at | 17419 | -0.116 | 0.0063 | No | ||
| 52 | GMNN | 1417506_at | 18961 | -0.168 | -0.0557 | No | ||
| 53 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 19008 | -0.170 | -0.0492 | No | ||
| 54 | DHRS2 | 1421663_at | 19165 | -0.177 | -0.0474 | No | ||
| 55 | AIF1 | 1418204_s_at | 19318 | -0.186 | -0.0449 | No | ||
| 56 | NOTCH2 | 1451889_at 1455556_at | 19560 | -0.201 | -0.0458 | No | ||
| 57 | TBRG1 | 1452648_at | 19887 | -0.226 | -0.0493 | No | ||
| 58 | CDK5RAP1 | 1448626_at | 19972 | -0.233 | -0.0414 | No | ||
| 59 | CDKN1C | 1417649_at | 19980 | -0.234 | -0.0299 | No | ||
| 60 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20040 | -0.240 | -0.0205 | No | ||
| 61 | MYC | 1424942_a_at | 20287 | -0.268 | -0.0182 | No | ||
| 62 | SESN1 | 1433711_s_at 1438931_s_at 1441649_at 1444376_at 1454699_at | 20345 | -0.275 | -0.0069 | No | ||
| 63 | RUNX3 | 1421467_at 1440275_at | 20351 | -0.276 | 0.0068 | No | ||
| 64 | KHDRBS1 | 1418628_at 1418630_at | 20463 | -0.290 | 0.0164 | No | ||
| 65 | TBRG4 | 1417658_at 1442793_s_at 1448795_a_at 1448796_s_at | 20467 | -0.291 | 0.0310 | No | ||
| 66 | APBB2 | 1426719_at 1426720_at 1440135_at 1446481_at 1452342_at | 20807 | -0.349 | 0.0331 | No | ||
| 67 | INHBA | 1422053_at 1458291_at | 20861 | -0.364 | 0.0491 | No |

