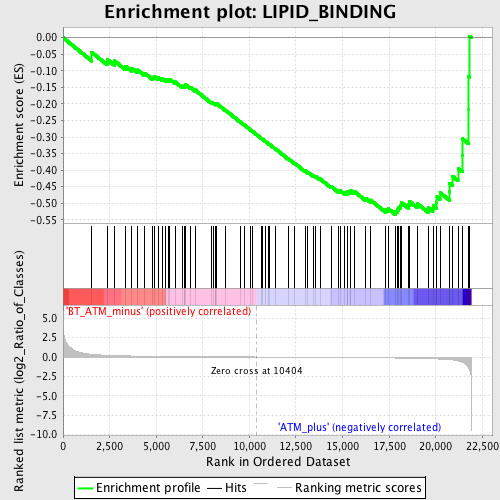
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | LIPID_BINDING |
| Enrichment Score (ES) | -0.5340335 |
| Normalized Enrichment Score (NES) | -1.5560042 |
| Nominal p-value | 0.018018018 |
| FDR q-value | 0.8055068 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 1542 | 0.345 | -0.0444 | No | ||
| 2 | SCIN | 1450276_a_at | 2367 | 0.216 | -0.0658 | No | ||
| 3 | PGRMC1 | 1423451_at | 2746 | 0.179 | -0.0695 | No | ||
| 4 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 3355 | 0.142 | -0.0866 | No | ||
| 5 | GPAA1 | 1438152_at 1448225_at 1455954_x_at | 3684 | 0.128 | -0.0920 | No | ||
| 6 | SDPR | 1416778_at 1416779_at 1443832_s_at | 3998 | 0.116 | -0.0975 | No | ||
| 7 | PLA2G7 | 1430700_a_at | 4388 | 0.103 | -0.1076 | No | ||
| 8 | SHC1 | 1422853_at 1422854_at | 4792 | 0.092 | -0.1190 | No | ||
| 9 | BAX | 1416837_at | 4885 | 0.090 | -0.1165 | No | ||
| 10 | HDLBP | 1415987_at 1415988_at 1419806_at 1419869_s_at 1443546_at 1449615_s_at | 5106 | 0.085 | -0.1201 | No | ||
| 11 | ANXA9 | 1431554_a_at | 5333 | 0.080 | -0.1244 | No | ||
| 12 | DOC2B | 1420666_at 1420667_at | 5489 | 0.076 | -0.1258 | No | ||
| 13 | PICALM | 1441328_at 1443218_at 1446968_at 1451316_a_at 1455773_at | 5637 | 0.073 | -0.1269 | No | ||
| 14 | RASAL1 | 1417858_at | 5726 | 0.071 | -0.1256 | No | ||
| 15 | ZFYVE16 | 1447947_at | 6011 | 0.066 | -0.1336 | No | ||
| 16 | EEA1 | 1438045_at | 6386 | 0.059 | -0.1463 | No | ||
| 17 | SYP | 1448280_at 1456249_x_at | 6500 | 0.057 | -0.1471 | No | ||
| 18 | PGRMC2 | 1428714_at 1452882_at | 6530 | 0.056 | -0.1442 | No | ||
| 19 | GC | 1426547_at | 6580 | 0.055 | -0.1422 | No | ||
| 20 | CENTD3 | 1419833_s_at 1447407_at 1451282_at | 6838 | 0.051 | -0.1501 | No | ||
| 21 | PCLO | 1419392_at 1452423_at | 7084 | 0.047 | -0.1578 | No | ||
| 22 | CAV1 | 1449145_a_at | 7955 | 0.033 | -0.1951 | No | ||
| 23 | ALDH6A1 | 1448104_at | 8060 | 0.032 | -0.1974 | No | ||
| 24 | PEBP1 | 1415950_a_at 1438649_x_at | 8192 | 0.030 | -0.2011 | No | ||
| 25 | APOA5 | 1417610_at | 8206 | 0.030 | -0.1995 | No | ||
| 26 | ACOT7 | 1417094_at | 8222 | 0.030 | -0.1979 | No | ||
| 27 | EDG3 | 1437173_at 1438658_a_at 1460661_at | 8727 | 0.022 | -0.2193 | No | ||
| 28 | MAL | 1417275_at 1432558_a_at | 9513 | 0.012 | -0.2543 | No | ||
| 29 | APOA1 | 1419232_a_at 1419233_x_at 1438840_x_at 1455201_x_at | 9729 | 0.009 | -0.2635 | No | ||
| 30 | WDFY1 | 1424749_at 1435588_at 1437358_at 1447543_at 1453426_a_at | 10047 | 0.005 | -0.2776 | No | ||
| 31 | THY1 | 1423135_at | 10173 | 0.003 | -0.2831 | No | ||
| 32 | EDG5 | 1428176_at 1447880_x_at | 10673 | -0.003 | -0.3057 | No | ||
| 33 | LYPLA3 | 1422341_s_at 1423704_at | 10715 | -0.004 | -0.3073 | No | ||
| 34 | SYTL4 | 1417336_a_at 1425998_at | 10850 | -0.006 | -0.3130 | No | ||
| 35 | MCTP1 | 1430439_at | 11054 | -0.009 | -0.3216 | No | ||
| 36 | PSAP | 1415687_a_at 1421813_a_at | 11097 | -0.009 | -0.3228 | No | ||
| 37 | STARD3 | 1417405_at | 11393 | -0.013 | -0.3354 | No | ||
| 38 | FABP7 | 1450779_at | 12079 | -0.021 | -0.3651 | No | ||
| 39 | ESR2 | 1426103_a_at | 12435 | -0.026 | -0.3793 | No | ||
| 40 | SYT1 | 1421990_at 1431191_a_at 1433884_at 1438282_at 1442557_at 1457497_at | 12993 | -0.034 | -0.4023 | No | ||
| 41 | APOA4 | 1417761_at 1436504_x_at | 13149 | -0.036 | -0.4066 | No | ||
| 42 | RASGRP4 | 1425380_at | 13449 | -0.041 | -0.4172 | No | ||
| 43 | FABP4 | 1417023_a_at 1424155_at 1425809_at 1451263_a_at | 13579 | -0.043 | -0.4199 | No | ||
| 44 | SHBG | 1421602_at | 13799 | -0.046 | -0.4265 | No | ||
| 45 | FABP2 | 1418438_at | 14385 | -0.055 | -0.4490 | No | ||
| 46 | EDG7 | 1418723_at | 14770 | -0.061 | -0.4620 | No | ||
| 47 | PAFAH2 | 1448489_at | 14876 | -0.063 | -0.4620 | No | ||
| 48 | CPNE3 | 1428514_at 1432021_at 1435450_at 1452814_at | 15120 | -0.067 | -0.4681 | No | ||
| 49 | FABP5 | 1416022_at | 15248 | -0.069 | -0.4687 | No | ||
| 50 | CENTD2 | 1428064_at | 15282 | -0.069 | -0.4649 | No | ||
| 51 | APOF | 1418239_at | 15414 | -0.072 | -0.4655 | No | ||
| 52 | CENTA2 | 1425638_at | 15448 | -0.073 | -0.4615 | No | ||
| 53 | APOL2 | 1441054_at | 15649 | -0.076 | -0.4649 | No | ||
| 54 | APOC2 | 1418069_at | 16260 | -0.088 | -0.4862 | No | ||
| 55 | NPC2 | 1416901_at 1443266_at 1448513_a_at 1455768_at | 16504 | -0.093 | -0.4903 | No | ||
| 56 | EDG4 | 1420576_at 1442291_at | 17328 | -0.113 | -0.5193 | No | ||
| 57 | GLTP | 1419027_s_at | 17454 | -0.117 | -0.5162 | No | ||
| 58 | ALDH1A1 | 1416468_at 1445980_at | 17845 | -0.128 | -0.5243 | Yes | ||
| 59 | RAB35 | 1433922_at | 17938 | -0.131 | -0.5187 | Yes | ||
| 60 | HSD17B4 | 1417369_at 1455777_x_at | 18032 | -0.134 | -0.5128 | Yes | ||
| 61 | CPNE1 | 1433439_at | 18111 | -0.136 | -0.5060 | Yes | ||
| 62 | TSPO | 1416695_at 1438948_x_at 1456251_x_at | 18150 | -0.137 | -0.4974 | Yes | ||
| 63 | ZFYVE1 | 1435071_at 1435072_at | 18564 | -0.152 | -0.5048 | Yes | ||
| 64 | PTAFR | 1427871_at 1427872_at | 18594 | -0.152 | -0.4946 | Yes | ||
| 65 | OSBP | 1460350_at | 19011 | -0.170 | -0.5008 | Yes | ||
| 66 | GRK5 | 1446361_at 1446998_at 1449514_at | 19607 | -0.203 | -0.5126 | Yes | ||
| 67 | CENTD1 | 1436895_at 1446649_at 1452291_at 1456337_at 1458173_at | 19864 | -0.223 | -0.5074 | Yes | ||
| 68 | ADH5 | 1416185_a_at 1443380_at | 20032 | -0.240 | -0.4969 | Yes | ||
| 69 | PLEKHA2 | 1417288_at 1417289_at | 20077 | -0.244 | -0.4805 | Yes | ||
| 70 | PLEKHA3 | 1420839_at 1420840_at 1447772_at | 20240 | -0.263 | -0.4680 | Yes | ||
| 71 | PIGK | 1428488_at 1435352_at 1452801_at | 20756 | -0.338 | -0.4660 | Yes | ||
| 72 | EDG6 | 1451024_at | 20761 | -0.339 | -0.4405 | Yes | ||
| 73 | APOE | 1432466_a_at | 20907 | -0.378 | -0.4185 | Yes | ||
| 74 | RASGRP1 | 1421176_at 1431749_a_at 1434295_at 1450143_at | 21207 | -0.490 | -0.3951 | Yes | ||
| 75 | PRKCI | 1417410_s_at 1448695_at | 21433 | -0.662 | -0.3553 | Yes | ||
| 76 | SCP2 | 1419974_at 1426219_at 1449686_s_at | 21438 | -0.667 | -0.3051 | Yes | ||
| 77 | AKR1D1 | 1425771_at 1455100_at | 21757 | -1.343 | -0.2180 | Yes | ||
| 78 | PLEKHA1 | 1417965_at 1443111_at | 21759 | -1.350 | -0.1159 | Yes | ||
| 79 | PSCD3 | 1418758_a_at 1431707_a_at 1446268_at | 21831 | -1.637 | 0.0047 | Yes |

