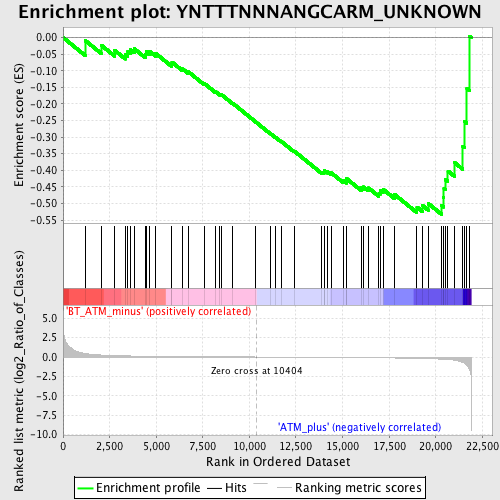
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | YNTTTNNNANGCARM_UNKNOWN |
| Enrichment Score (ES) | -0.5332622 |
| Normalized Enrichment Score (NES) | -1.5029826 |
| Nominal p-value | 0.03021148 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.893 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GPR158 | 1438526_at 1459430_at | 1186 | 0.459 | -0.0088 | No | ||
| 2 | LDB2 | 1421101_a_at 1439557_s_at 1445126_at 1456786_at | 2059 | 0.252 | -0.0237 | No | ||
| 3 | KCNA2 | 1422197_at | 2769 | 0.177 | -0.0385 | No | ||
| 4 | EVX1 | 1421627_at | 3344 | 0.143 | -0.0507 | No | ||
| 5 | GABRG1 | 1427227_at 1460408_at | 3478 | 0.136 | -0.0432 | No | ||
| 6 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 3607 | 0.131 | -0.0361 | No | ||
| 7 | ELAVL2 | 1421881_a_at 1421882_a_at 1421883_at 1433052_at | 3821 | 0.122 | -0.0337 | No | ||
| 8 | CDX2 | 1422074_at 1422075_at | 4432 | 0.101 | -0.0516 | No | ||
| 9 | SFRP2 | 1448201_at | 4455 | 0.101 | -0.0426 | No | ||
| 10 | LRRTM1 | 1437746_at 1452624_at 1455883_a_at | 4637 | 0.096 | -0.0413 | No | ||
| 11 | GNAI1 | 1427510_at 1434440_at 1454959_s_at | 4970 | 0.088 | -0.0478 | No | ||
| 12 | HOXC4 | 1422870_at | 5840 | 0.069 | -0.0807 | No | ||
| 13 | TLL2 | 1421526_at | 5844 | 0.069 | -0.0740 | No | ||
| 14 | HOXB6 | 1451660_a_at | 6419 | 0.058 | -0.0945 | No | ||
| 15 | DNAH5 | 1421434_at | 6706 | 0.053 | -0.1023 | No | ||
| 16 | C1S | 1424041_s_at | 7569 | 0.039 | -0.1378 | No | ||
| 17 | KCTD12 | 1434881_s_at | 8201 | 0.030 | -0.1637 | No | ||
| 18 | FGF14 | 1435747_at 1447221_x_at 1447480_at 1449958_a_at 1458830_at 1459073_x_at | 8415 | 0.027 | -0.1708 | No | ||
| 19 | IL10 | 1450330_at | 8488 | 0.026 | -0.1715 | No | ||
| 20 | NRK | 1436399_s_at 1450078_at 1450079_at | 9105 | 0.018 | -0.1979 | No | ||
| 21 | B3GNT7 | 1436321_at | 10346 | 0.001 | -0.2545 | No | ||
| 22 | FGD4 | 1425037_at 1426041_a_at 1426042_at 1451659_at 1455337_at | 11135 | -0.010 | -0.2896 | No | ||
| 23 | SMPX | 1418095_at | 11396 | -0.013 | -0.3002 | No | ||
| 24 | LEMD2 | 1424326_at | 11719 | -0.017 | -0.3132 | No | ||
| 25 | ZFHX1B | 1422748_at 1434298_at 1442393_at 1445518_at 1454200_at 1456389_at | 12414 | -0.026 | -0.3423 | No | ||
| 26 | KIF13A | 1439532_s_at 1440578_at 1442252_at 1446026_at 1447853_x_at 1451890_at 1455746_at | 13898 | -0.047 | -0.4054 | No | ||
| 27 | VDR | 1418175_at 1418176_at | 14011 | -0.049 | -0.4057 | No | ||
| 28 | FGF6 | 1427582_at | 14013 | -0.049 | -0.4008 | No | ||
| 29 | ADAMTS5 | 1422561_at 1450658_at 1456404_at | 14180 | -0.052 | -0.4033 | No | ||
| 30 | GCDH | 1448717_at | 14403 | -0.056 | -0.4080 | No | ||
| 31 | FGF12 | 1440270_at 1451693_a_at 1460137_at | 15042 | -0.066 | -0.4306 | No | ||
| 32 | RSN | 1425060_s_at 1431098_at 1439800_at 1444750_at 1450351_a_at 1452929_at | 15219 | -0.068 | -0.4319 | No | ||
| 33 | EYA1 | 1421727_at 1443117_at 1457424_at | 15225 | -0.069 | -0.4253 | No | ||
| 34 | OMG | 1418212_at | 16008 | -0.083 | -0.4529 | No | ||
| 35 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 16135 | -0.085 | -0.4502 | No | ||
| 36 | GPM6A | 1426442_at 1456741_s_at 1459303_at | 16400 | -0.091 | -0.4533 | No | ||
| 37 | VAX1 | 1421774_at | 16961 | -0.104 | -0.4686 | No | ||
| 38 | HCRTR2 | 1444223_at | 17031 | -0.106 | -0.4612 | No | ||
| 39 | HOXC5 | 1439885_at 1450832_at 1456301_at | 17184 | -0.110 | -0.4573 | No | ||
| 40 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 17794 | -0.126 | -0.4726 | No | ||
| 41 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 19002 | -0.170 | -0.5110 | Yes | ||
| 42 | PVRL1 | 1438111_at 1438421_at 1440131_at 1446488_at 1450819_at | 19290 | -0.184 | -0.5058 | Yes | ||
| 43 | GRK5 | 1446361_at 1446998_at 1449514_at | 19607 | -0.203 | -0.5001 | Yes | ||
| 44 | MSX2 | 1438351_at 1449559_at | 20333 | -0.274 | -0.5061 | Yes | ||
| 45 | KLHL5 | 1426530_a_at 1444486_at 1449602_at | 20438 | -0.288 | -0.4824 | Yes | ||
| 46 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 20451 | -0.290 | -0.4542 | Yes | ||
| 47 | MAFB | 1451715_at 1451716_at | 20525 | -0.300 | -0.4279 | Yes | ||
| 48 | GABRA1 | 1421280_at 1421281_at 1436889_at 1455766_at | 20667 | -0.324 | -0.4023 | Yes | ||
| 49 | PDE4D | 1443289_at 1458212_at 1459289_at 1459311_at | 21035 | -0.420 | -0.3775 | Yes | ||
| 50 | CNN3 | 1426724_at 1436759_x_at 1436836_x_at 1438354_x_at 1455570_x_at 1456380_x_at | 21466 | -0.695 | -0.3283 | Yes | ||
| 51 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 21554 | -0.797 | -0.2533 | Yes | ||
| 52 | NRF1 | 1420978_at 1424787_a_at 1434627_at 1439545_at 1445914_at | 21685 | -1.070 | -0.1533 | Yes | ||
| 53 | SYNPR | 1423640_at 1444331_at | 21840 | -1.662 | 0.0043 | Yes |

