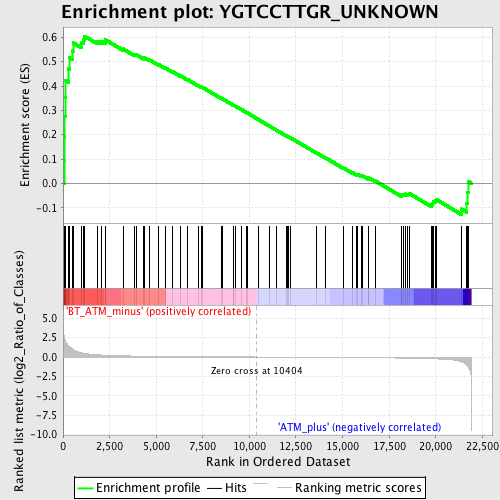
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | YGTCCTTGR_UNKNOWN |
| Enrichment Score (ES) | 0.6051397 |
| Normalized Enrichment Score (NES) | 1.594772 |
| Nominal p-value | 0.0074294205 |
| FDR q-value | 0.27754697 |
| FWER p-Value | 0.761 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CITED2 | 1417129_a_at 1421267_a_at 1439915_at 1440091_at 1443561_at 1443926_at 1445638_at 1446176_at 1447861_x_at 1457632_s_at 1459301_at | 54 | 2.704 | 0.0959 | Yes | ||
| 2 | SLC38A2 | 1426722_at 1429593_at 1429594_at | 64 | 2.633 | 0.1914 | Yes | ||
| 3 | CYCS | 1422484_at 1445484_at | 92 | 2.381 | 0.2768 | Yes | ||
| 4 | PHYHIPL | 1427023_at 1437062_s_at | 114 | 2.135 | 0.3535 | Yes | ||
| 5 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 147 | 1.927 | 0.4222 | Yes | ||
| 6 | PPP2R4 | 1439383_x_at 1439393_x_at 1448138_at | 282 | 1.497 | 0.4705 | Yes | ||
| 7 | HMGCS1 | 1433443_a_at 1433444_at 1433445_x_at 1433446_at 1441536_at | 346 | 1.339 | 0.5164 | Yes | ||
| 8 | CXXC4 | 1437351_at 1438304_at 1456811_at | 513 | 0.981 | 0.5445 | Yes | ||
| 9 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 534 | 0.952 | 0.5782 | Yes | ||
| 10 | H2AFZ | 1441370_at | 992 | 0.541 | 0.5770 | Yes | ||
| 11 | PANK3 | 1426259_at | 1086 | 0.495 | 0.5908 | Yes | ||
| 12 | PAM | 1418908_at 1445797_at | 1148 | 0.470 | 0.6051 | Yes | ||
| 13 | CHD6 | 1427384_at 1431047_at | 1834 | 0.288 | 0.5843 | No | ||
| 14 | AMY2A | 1457367_at | 2052 | 0.253 | 0.5836 | No | ||
| 15 | SCN2B | 1430648_at 1436134_at 1457144_at | 2265 | 0.227 | 0.5821 | No | ||
| 16 | LDHB | 1416183_a_at 1434499_a_at 1442618_at 1448237_x_at 1455235_x_at | 2268 | 0.226 | 0.5903 | No | ||
| 17 | BAZ2A | 1427209_at 1427210_at 1438192_s_at | 3215 | 0.149 | 0.5524 | No | ||
| 18 | ELAVL2 | 1421881_a_at 1421882_a_at 1421883_at 1433052_at | 3821 | 0.122 | 0.5292 | No | ||
| 19 | VGF | 1436094_at | 3939 | 0.118 | 0.5281 | No | ||
| 20 | KCNJ16 | 1435094_at 1436163_at | 4317 | 0.105 | 0.5147 | No | ||
| 21 | GAS6 | 1417399_at 1459585_at | 4362 | 0.103 | 0.5165 | No | ||
| 22 | SYT13 | 1434470_at 1451045_at | 4645 | 0.096 | 0.5071 | No | ||
| 23 | SOCS5 | 1423349_at 1423350_at 1441640_at 1442890_at | 5140 | 0.084 | 0.4875 | No | ||
| 24 | CLCNKA | 1450182_at | 5490 | 0.076 | 0.4743 | No | ||
| 25 | ZIC3 | 1423424_at 1438737_at | 5867 | 0.068 | 0.4596 | No | ||
| 26 | CTRL | 1431763_a_at | 6307 | 0.060 | 0.4417 | No | ||
| 27 | HRH3 | 1445285_at 1448807_at | 6682 | 0.054 | 0.4266 | No | ||
| 28 | GABRA3 | 1421263_at | 7259 | 0.044 | 0.4018 | No | ||
| 29 | MCAM | 1416357_a_at | 7419 | 0.041 | 0.3961 | No | ||
| 30 | TMEM16D | 1442143_at | 7504 | 0.040 | 0.3937 | No | ||
| 31 | NPTX2 | 1420720_at 1449960_at | 8528 | 0.025 | 0.3478 | No | ||
| 32 | AAK1 | 1420026_at 1435038_s_at 1441782_at | 8560 | 0.025 | 0.3473 | No | ||
| 33 | IGSF3 | 1431322_at 1455048_at 1455049_at | 9147 | 0.017 | 0.3211 | No | ||
| 34 | NEF3 | 1422520_at | 9170 | 0.017 | 0.3207 | No | ||
| 35 | SNCB | 1418053_at | 9265 | 0.015 | 0.3170 | No | ||
| 36 | MT3 | 1420575_at | 9578 | 0.011 | 0.3031 | No | ||
| 37 | WBSCR1 | 1416941_s_at 1436637_at 1438554_x_at 1442942_at 1447070_at 1451521_x_at | 9859 | 0.007 | 0.2906 | No | ||
| 38 | SEMA6D | 1446536_at 1448060_at 1453055_at 1459318_at | 9911 | 0.007 | 0.2885 | No | ||
| 39 | CRAT | 1417008_at 1441919_x_at 1448544_at | 10496 | -0.001 | 0.2618 | No | ||
| 40 | FHL2 | 1419184_a_at | 11062 | -0.009 | 0.2363 | No | ||
| 41 | ATP5B | 1416829_at | 11453 | -0.014 | 0.2189 | No | ||
| 42 | TRIM2 | 1417027_at 1417028_a_at 1417029_a_at 1441487_at 1448551_a_at 1459860_x_at | 11986 | -0.020 | 0.1953 | No | ||
| 43 | HDAC3 | 1426437_s_at | 12043 | -0.021 | 0.1935 | No | ||
| 44 | OTX2 | 1425926_a_at | 12127 | -0.022 | 0.1906 | No | ||
| 45 | RGL1 | 1418535_at 1440662_at 1449124_at | 12222 | -0.024 | 0.1871 | No | ||
| 46 | DRD3 | 1422278_at | 13609 | -0.043 | 0.1253 | No | ||
| 47 | SNTA1 | 1416825_at | 14082 | -0.050 | 0.1055 | No | ||
| 48 | KCNAB3 | 1421790_a_at 1423950_at | 15076 | -0.066 | 0.0625 | No | ||
| 49 | ADAM11 | 1450248_at | 15548 | -0.074 | 0.0437 | No | ||
| 50 | EMX2 | 1456258_at | 15754 | -0.078 | 0.0371 | No | ||
| 51 | HOXD11 | 1450584_at | 15802 | -0.079 | 0.0378 | No | ||
| 52 | ARMET | 1428112_at 1459455_at | 16007 | -0.083 | 0.0315 | No | ||
| 53 | THRA | 1426997_at 1443952_at 1454675_at | 16064 | -0.084 | 0.0320 | No | ||
| 54 | CHST8 | 1428977_at | 16390 | -0.091 | 0.0204 | No | ||
| 55 | SYT7 | 1423012_at 1441927_at 1446641_at 1460081_at | 16398 | -0.091 | 0.0234 | No | ||
| 56 | CS | 1422577_at 1422578_at 1450667_a_at | 16762 | -0.099 | 0.0104 | No | ||
| 57 | CBX4 | 1419583_at 1440479_at | 18183 | -0.138 | -0.0495 | No | ||
| 58 | PRKACA | 1447720_x_at 1450519_a_at | 18194 | -0.139 | -0.0449 | No | ||
| 59 | STIM1 | 1436945_x_at 1448320_at 1458460_at | 18293 | -0.142 | -0.0442 | No | ||
| 60 | AFP | 1416645_a_at 1416646_at 1436879_x_at 1436880_at 1436881_x_at | 18394 | -0.145 | -0.0435 | No | ||
| 61 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 18519 | -0.150 | -0.0437 | No | ||
| 62 | TNKS1BP1 | 1427051_at | 18603 | -0.153 | -0.0420 | No | ||
| 63 | HSPBAP1 | 1428566_at 1428567_at 1441900_x_at 1442842_at | 19775 | -0.216 | -0.0877 | No | ||
| 64 | VAMP2 | 1420833_at 1420834_at | 19854 | -0.222 | -0.0832 | No | ||
| 65 | CENTD1 | 1436895_at 1446649_at 1452291_at 1456337_at 1458173_at | 19864 | -0.223 | -0.0755 | No | ||
| 66 | PRKAG1 | 1417690_at 1433533_x_at 1457803_at | 19974 | -0.233 | -0.0719 | No | ||
| 67 | PRKCE | 1437860_at 1437861_s_at 1442609_at 1444649_at 1449956_at 1452878_at | 20057 | -0.241 | -0.0669 | No | ||
| 68 | ARPC5 | 1444568_at 1448129_at | 21388 | -0.623 | -0.1051 | No | ||
| 69 | DIRC2 | 1454654_at 1457245_at | 21671 | -1.024 | -0.0807 | No | ||
| 70 | ID2 | 1422537_a_at 1435176_a_at 1453596_at | 21727 | -1.235 | -0.0383 | No | ||
| 71 | SYNPO2L | 1428295_at 1447657_s_at 1447658_x_at 1458691_at | 21748 | -1.311 | 0.0085 | No |

