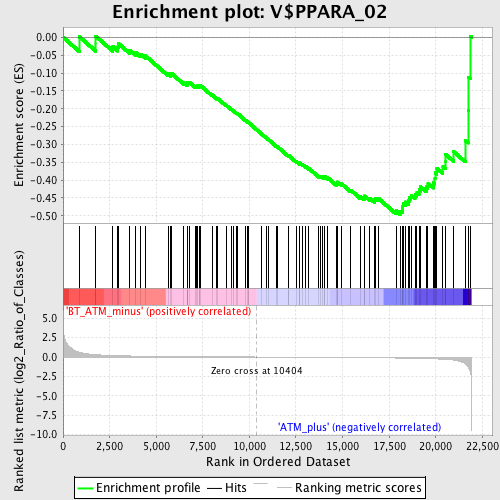
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$PPARA_02 |
| Enrichment Score (ES) | -0.4962129 |
| Normalized Enrichment Score (NES) | -1.5476575 |
| Nominal p-value | 0.009933775 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.786 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SUPT16H | 1419741_at 1449578_at 1456449_at | 875 | 0.606 | 0.0015 | No | ||
| 2 | CBX6 | 1424407_s_at 1429290_at | 1753 | 0.304 | -0.0178 | No | ||
| 3 | HOXA9 | 1421579_at 1455626_at | 1754 | 0.303 | 0.0031 | No | ||
| 4 | PERQ1 | 1423460_at | 2674 | 0.185 | -0.0263 | No | ||
| 5 | SMARCA5 | 1440048_at | 2940 | 0.165 | -0.0271 | No | ||
| 6 | ADD3 | 1423297_at 1423298_at 1426574_a_at 1447077_at 1456162_x_at | 2986 | 0.163 | -0.0180 | No | ||
| 7 | ZSWIM3 | 1428313_at 1444571_at 1457785_at | 3580 | 0.132 | -0.0361 | No | ||
| 8 | NEUROG1 | 1438551_at 1450836_at | 3893 | 0.120 | -0.0422 | No | ||
| 9 | ARHGAP26 | 1430957_at 1435694_at 1440688_at | 4161 | 0.109 | -0.0469 | No | ||
| 10 | POGZ | 1434650_at 1454827_at 1455046_a_at 1457748_at | 4422 | 0.102 | -0.0518 | No | ||
| 11 | NEUROD2 | 1444362_at | 5636 | 0.073 | -0.1023 | No | ||
| 12 | RIN1 | 1424507_at 1437460_x_at | 5778 | 0.070 | -0.1040 | No | ||
| 13 | LMO1 | 1418478_at | 5806 | 0.070 | -0.1004 | No | ||
| 14 | GGN | 1445278_at | 6487 | 0.057 | -0.1276 | No | ||
| 15 | PPP1R9B | 1447247_at 1447486_at | 6657 | 0.054 | -0.1317 | No | ||
| 16 | HRH3 | 1445285_at 1448807_at | 6682 | 0.054 | -0.1291 | No | ||
| 17 | CHD2 | 1444246_at 1445843_at | 6687 | 0.054 | -0.1256 | No | ||
| 18 | CACNA1S | 1420442_at | 6795 | 0.052 | -0.1269 | No | ||
| 19 | VEGF | 1420909_at 1451959_a_at | 7101 | 0.047 | -0.1377 | No | ||
| 20 | ATP5G2 | 1415980_at 1456128_at | 7170 | 0.045 | -0.1377 | No | ||
| 21 | CYC1 | 1416604_at | 7206 | 0.045 | -0.1362 | No | ||
| 22 | WDR20 | 1430004_s_at 1453693_at | 7301 | 0.043 | -0.1375 | No | ||
| 23 | SLITRK5 | 1429718_at | 7323 | 0.043 | -0.1356 | No | ||
| 24 | EPN1 | 1427039_at | 7382 | 0.042 | -0.1354 | No | ||
| 25 | TSNAXIP1 | 1427018_at | 7998 | 0.033 | -0.1613 | No | ||
| 26 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 8254 | 0.029 | -0.1709 | No | ||
| 27 | KIF1C | 1424747_at 1426325_at | 8310 | 0.028 | -0.1715 | No | ||
| 28 | NXPH3 | 1419710_at | 8750 | 0.022 | -0.1901 | No | ||
| 29 | SEMA3A | 1420416_at 1420417_at 1449865_at | 9035 | 0.018 | -0.2018 | No | ||
| 30 | KRTAP6-3 | 1427842_at 1452575_at | 9151 | 0.017 | -0.2059 | No | ||
| 31 | RTN2 | 1419056_at | 9319 | 0.015 | -0.2126 | No | ||
| 32 | CTDSP1 | 1452055_at | 9338 | 0.014 | -0.2125 | No | ||
| 33 | ESRRG | 1421747_at 1455267_at 1457896_at | 9793 | 0.008 | -0.2327 | No | ||
| 34 | CDH17 | 1419331_at | 9896 | 0.007 | -0.2369 | No | ||
| 35 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 9944 | 0.006 | -0.2386 | No | ||
| 36 | PSCA | 1451258_at | 9968 | 0.006 | -0.2393 | No | ||
| 37 | SERPINF2 | 1417498_at | 10668 | -0.003 | -0.2711 | No | ||
| 38 | ABL1 | 1423999_at 1441291_at 1444134_at 1445153_at | 10904 | -0.007 | -0.2814 | No | ||
| 39 | ROM1 | 1448996_at | 11006 | -0.008 | -0.2854 | No | ||
| 40 | TRPV6 | 1419615_at | 11479 | -0.014 | -0.3061 | No | ||
| 41 | KCNB1 | 1423179_at 1423180_at | 11523 | -0.015 | -0.3071 | No | ||
| 42 | HR | 1422110_at 1435950_at | 12120 | -0.022 | -0.3328 | No | ||
| 43 | TNXB | 1450798_at | 12123 | -0.022 | -0.3314 | No | ||
| 44 | WNT4 | 1450782_at | 12549 | -0.028 | -0.3489 | No | ||
| 45 | OTX1 | 1437601_at | 12695 | -0.030 | -0.3535 | No | ||
| 46 | NLGN3 | 1456384_at | 12703 | -0.030 | -0.3518 | No | ||
| 47 | HMGN2 | 1433507_a_at | 12840 | -0.032 | -0.3558 | No | ||
| 48 | EPHB1 | 1455188_at | 13030 | -0.034 | -0.3621 | No | ||
| 49 | MAP3K11 | 1450669_at | 13197 | -0.037 | -0.3672 | No | ||
| 50 | ASB7 | 1435010_at 1438744_at 1446319_at 1450363_at | 13733 | -0.045 | -0.3886 | No | ||
| 51 | PGF | 1418471_at | 13841 | -0.046 | -0.3903 | No | ||
| 52 | PHKG2 | 1428511_at | 13923 | -0.048 | -0.3908 | No | ||
| 53 | FGF6 | 1427582_at | 14013 | -0.049 | -0.3915 | No | ||
| 54 | PRX | 1423292_a_at | 14041 | -0.049 | -0.3893 | No | ||
| 55 | LAMA5 | 1459614_at | 14211 | -0.052 | -0.3935 | No | ||
| 56 | UCN2 | 1450607_s_at | 14684 | -0.060 | -0.4110 | No | ||
| 57 | FSCN3 | 1418948_at | 14717 | -0.061 | -0.4083 | No | ||
| 58 | YARS | 1460638_at | 14742 | -0.061 | -0.4052 | No | ||
| 59 | CDK5R2 | 1450465_at | 14939 | -0.064 | -0.4098 | No | ||
| 60 | CENTA2 | 1425638_at | 15448 | -0.073 | -0.4280 | No | ||
| 61 | PHF12 | 1434922_at 1453579_at | 15961 | -0.082 | -0.4458 | No | ||
| 62 | MAN2B1 | 1416340_a_at 1436781_at | 16166 | -0.086 | -0.4493 | No | ||
| 63 | PKP3 | 1418831_at 1418832_at | 16189 | -0.086 | -0.4444 | No | ||
| 64 | DHH | 1422127_at 1434959_at | 16473 | -0.092 | -0.4510 | No | ||
| 65 | LOXL4 | 1421153_at 1450134_at | 16718 | -0.098 | -0.4555 | No | ||
| 66 | PDGFB | 1450413_at 1450414_at | 16775 | -0.099 | -0.4512 | No | ||
| 67 | NKIRAS2 | 1424416_at | 16930 | -0.103 | -0.4512 | No | ||
| 68 | MSI1 | 1421409_at 1444306_at | 17879 | -0.129 | -0.4857 | No | ||
| 69 | SCAMP3 | 1416294_at | 18109 | -0.136 | -0.4869 | Yes | ||
| 70 | OSM | 1438767_at | 18218 | -0.139 | -0.4822 | Yes | ||
| 71 | AIM1L | 1437813_at | 18229 | -0.140 | -0.4731 | Yes | ||
| 72 | PAK1 | 1420979_at 1420980_at 1450070_s_at 1459029_at | 18252 | -0.140 | -0.4645 | Yes | ||
| 73 | DNAJC7 | 1448362_at 1459704_at | 18399 | -0.145 | -0.4612 | Yes | ||
| 74 | RAD23B | 1424222_s_at 1450903_at 1455420_at 1456822_at | 18541 | -0.151 | -0.4573 | Yes | ||
| 75 | DCTN1 | 1422521_at 1435414_s_at | 18578 | -0.152 | -0.4485 | Yes | ||
| 76 | POFUT1 | 1443881_at 1450137_at 1456323_at | 18681 | -0.156 | -0.4425 | Yes | ||
| 77 | RGS8 | 1453060_at | 18898 | -0.165 | -0.4410 | Yes | ||
| 78 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 19002 | -0.170 | -0.4341 | Yes | ||
| 79 | CREB3L2 | 1437396_at 1442080_at 1452381_at | 19115 | -0.175 | -0.4272 | Yes | ||
| 80 | MNT | 1418192_at | 19190 | -0.179 | -0.4184 | Yes | ||
| 81 | RFX1 | 1422109_at 1436059_at | 19498 | -0.197 | -0.4189 | Yes | ||
| 82 | TITF1 | 1422346_at | 19593 | -0.203 | -0.4093 | Yes | ||
| 83 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 19874 | -0.224 | -0.4067 | Yes | ||
| 84 | BAHD1 | 1433703_s_at | 19968 | -0.233 | -0.3950 | Yes | ||
| 85 | PRKAG1 | 1417690_at 1433533_x_at 1457803_at | 19974 | -0.233 | -0.3792 | Yes | ||
| 86 | KPNB1 | 1416925_at 1434357_a_at 1448526_at 1451967_x_at | 20073 | -0.244 | -0.3670 | Yes | ||
| 87 | HOXA5 | 1443803_x_at 1448926_at | 20398 | -0.283 | -0.3624 | Yes | ||
| 88 | NRGN | 1423231_at | 20520 | -0.299 | -0.3474 | Yes | ||
| 89 | MAFB | 1451715_at 1451716_at | 20525 | -0.300 | -0.3270 | Yes | ||
| 90 | HIP2 | 1417186_at 1417187_at 1417188_s_at 1441012_at 1442671_at 1443148_at 1444212_at | 20968 | -0.394 | -0.3202 | Yes | ||
| 91 | FTS | 1423364_a_at 1430219_at 1437767_s_at | 21603 | -0.874 | -0.2892 | Yes | ||
| 92 | RTN4 | 1421116_a_at 1435284_at 1437224_at 1439650_at 1442760_x_at 1452649_at | 21752 | -1.330 | -0.2047 | Yes | ||
| 93 | ERF | 1422114_at 1435561_at | 21763 | -1.358 | -0.1119 | Yes | ||
| 94 | TRIB1 | 1424880_at 1424881_at | 21861 | -1.744 | 0.0033 | Yes |

