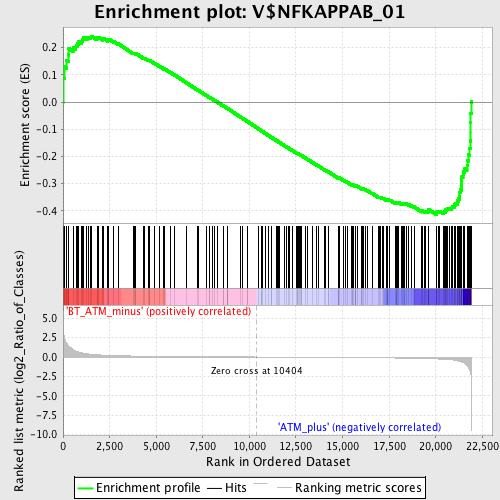
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$NFKAPPAB_01 |
| Enrichment Score (ES) | -0.41311553 |
| Normalized Enrichment Score (NES) | -1.3744501 |
| Nominal p-value | 0.024291499 |
| FDR q-value | 0.6712995 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 2 | 5.413 | 0.0890 | No | ||
| 2 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 58 | 2.672 | 0.1305 | No | ||
| 3 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 192 | 1.767 | 0.1534 | No | ||
| 4 | RBMS1 | 1418703_at 1434005_at 1441437_at 1458578_at 1459968_at | 286 | 1.485 | 0.1736 | No | ||
| 5 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 307 | 1.436 | 0.1963 | No | ||
| 6 | ZBTB11 | 1454826_at | 533 | 0.952 | 0.2016 | No | ||
| 7 | P2RY10 | 1452815_at | 691 | 0.757 | 0.2069 | No | ||
| 8 | HSD3B7 | 1416968_a_at 1442912_at | 774 | 0.678 | 0.2143 | No | ||
| 9 | OPCML | 1435612_at 1442752_at 1457446_at 1458228_at | 841 | 0.635 | 0.2217 | No | ||
| 10 | NR2F2 | 1416158_at 1416159_at 1416160_at 1431237_at 1444229_at | 989 | 0.542 | 0.2239 | No | ||
| 11 | IER5 | 1417612_at 1417613_at | 1053 | 0.511 | 0.2294 | No | ||
| 12 | CREB1 | 1421582_a_at 1421583_at 1423402_at 1428755_at 1452529_a_at 1452901_at | 1067 | 0.504 | 0.2371 | No | ||
| 13 | SP6 | 1427786_at 1427787_at 1437788_at 1442342_at | 1237 | 0.439 | 0.2365 | No | ||
| 14 | ZFP36L2 | 1437626_at 1439054_at 1451034_at | 1343 | 0.396 | 0.2382 | No | ||
| 15 | MOBKL2C | 1440919_at 1459919_a_at | 1459 | 0.362 | 0.2389 | No | ||
| 16 | RAP2C | 1428119_a_at 1437016_x_at 1460430_at | 1511 | 0.351 | 0.2423 | No | ||
| 17 | UBE2I | 1422712_a_at 1422713_a_at 1425478_x_at 1453189_at | 1821 | 0.290 | 0.2329 | No | ||
| 18 | CHD6 | 1427384_at 1431047_at | 1834 | 0.288 | 0.2371 | No | ||
| 19 | BMP2K | 1421103_at 1437419_at 1458370_at | 1901 | 0.276 | 0.2386 | No | ||
| 20 | ASH1L | 1420985_at 1441117_at 1450071_at 1450072_at | 2136 | 0.243 | 0.2318 | No | ||
| 21 | KCNS3 | 1445519_at 1457799_at | 2151 | 0.240 | 0.2352 | No | ||
| 22 | ITGB4 | 1427387_a_at | 2403 | 0.212 | 0.2271 | No | ||
| 23 | IL2RA | 1420691_at 1420692_at | 2426 | 0.209 | 0.2295 | No | ||
| 24 | ARPC2 | 1437148_at 1442295_at | 2460 | 0.206 | 0.2314 | No | ||
| 25 | PPARBP | 1421906_at 1421907_at 1439408_a_at 1446250_at 1447450_at 1448708_at 1450402_at 1453972_x_at | 2731 | 0.180 | 0.2220 | No | ||
| 26 | COL11A2 | 1423578_at | 2963 | 0.164 | 0.2140 | No | ||
| 27 | FBXL12 | 1448420_a_at | 3754 | 0.125 | 0.1798 | No | ||
| 28 | CCL5 | 1418126_at | 3811 | 0.123 | 0.1792 | No | ||
| 29 | LAMA1 | 1418153_at | 3879 | 0.120 | 0.1781 | No | ||
| 30 | FUT7 | 1420756_at | 3911 | 0.119 | 0.1786 | No | ||
| 31 | IFNB1 | 1422305_at | 4333 | 0.104 | 0.1610 | No | ||
| 32 | CHX10 | 1419628_at | 4392 | 0.103 | 0.1600 | No | ||
| 33 | HOXB9 | 1452317_at | 4585 | 0.098 | 0.1528 | No | ||
| 34 | DDR1 | 1415797_at 1415798_at 1435820_x_at 1437619_x_at 1438367_x_at 1439382_x_at 1456226_x_at 1459990_at | 4623 | 0.097 | 0.1527 | No | ||
| 35 | CLCN1 | 1427591_at 1448082_at | 4656 | 0.096 | 0.1528 | No | ||
| 36 | CTGF | 1416953_at | 4924 | 0.089 | 0.1420 | No | ||
| 37 | SLC6A12 | 1449382_at | 5163 | 0.083 | 0.1324 | No | ||
| 38 | BCKDK | 1443813_x_at 1460644_at | 5414 | 0.078 | 0.1222 | No | ||
| 39 | UACA | 1452985_at | 5454 | 0.077 | 0.1216 | No | ||
| 40 | CYLD | 1429617_at 1429618_at 1445213_at | 5742 | 0.071 | 0.1096 | No | ||
| 41 | NFKBIB | 1421266_s_at 1446718_at | 6000 | 0.066 | 0.0989 | No | ||
| 42 | PLAU | 1422138_at 1422139_at | 6605 | 0.055 | 0.0720 | No | ||
| 43 | IL1RAPL1 | 1445821_at 1458964_at | 7191 | 0.045 | 0.0459 | No | ||
| 44 | FGF1 | 1423136_at 1441042_at 1450869_at | 7291 | 0.043 | 0.0420 | No | ||
| 45 | PUNC | 1423496_a_at 1439498_at 1444981_at | 7676 | 0.038 | 0.0250 | No | ||
| 46 | AGPAT1 | 1421024_at 1421025_at | 7844 | 0.035 | 0.0179 | No | ||
| 47 | FGF17 | 1421523_at 1456239_at | 7874 | 0.035 | 0.0171 | No | ||
| 48 | SEC14L2 | 1424530_at | 8019 | 0.033 | 0.0110 | No | ||
| 49 | KCNH3 | 1421619_at 1431690_at 1445805_x_at 1459107_at | 8147 | 0.031 | 0.0057 | No | ||
| 50 | CXCL5 | 1419728_at | 8272 | 0.029 | 0.0005 | No | ||
| 51 | IL23A | 1419529_at | 8590 | 0.024 | -0.0137 | No | ||
| 52 | NFAT5 | 1426029_a_at 1438999_a_at 1439328_at 1439805_at 1448966_a_at 1451921_a_at 1458213_at | 8591 | 0.024 | -0.0133 | No | ||
| 53 | TSLP | 1450004_at | 8828 | 0.021 | -0.0238 | No | ||
| 54 | ARHGAP8 | 1451463_at | 8846 | 0.021 | -0.0243 | No | ||
| 55 | SMPD3 | 1422779_at 1438665_at 1445759_at 1450748_at | 8848 | 0.021 | -0.0240 | No | ||
| 56 | EIF5A | 1437859_x_at 1451470_s_at | 9520 | 0.012 | -0.0546 | No | ||
| 57 | ATOH1 | 1449822_at | 9611 | 0.011 | -0.0586 | No | ||
| 58 | SAMSN1 | 1421457_a_at 1431539_at | 9886 | 0.007 | -0.0711 | No | ||
| 59 | ACTN3 | 1418677_at | 9890 | 0.007 | -0.0711 | No | ||
| 60 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 10467 | -0.001 | -0.0976 | No | ||
| 61 | TRIM47 | 1426784_at | 10654 | -0.003 | -0.1061 | No | ||
| 62 | EDG5 | 1428176_at 1447880_x_at | 10673 | -0.003 | -0.1068 | No | ||
| 63 | AP1S2 | 1437998_at 1447903_x_at 1452657_at 1456252_x_at 1460036_at | 10696 | -0.004 | -0.1078 | No | ||
| 64 | RND1 | 1455197_at | 10875 | -0.006 | -0.1159 | No | ||
| 65 | SDC4 | 1417654_at 1448793_a_at | 11024 | -0.008 | -0.1225 | No | ||
| 66 | SLC12A2 | 1417622_at 1417623_at 1443332_at 1448780_at | 11192 | -0.010 | -0.1301 | No | ||
| 67 | BNC2 | 1438861_at 1440593_at 1459132_at | 11214 | -0.011 | -0.1308 | No | ||
| 68 | EGF | 1418093_a_at | 11481 | -0.014 | -0.1428 | No | ||
| 69 | WNT10B | 1426091_a_at | 11487 | -0.014 | -0.1428 | No | ||
| 70 | IL1A | 1421473_at | 11581 | -0.015 | -0.1469 | No | ||
| 71 | RIN2 | 1426368_at 1441209_at 1442829_at | 11632 | -0.016 | -0.1489 | No | ||
| 72 | KRTAP13-1 | 1428007_at | 11889 | -0.019 | -0.1604 | No | ||
| 73 | SALL1 | 1450489_at | 12002 | -0.020 | -0.1652 | No | ||
| 74 | NOL4 | 1430189_at 1441447_at 1456387_at | 12081 | -0.021 | -0.1684 | No | ||
| 75 | CPD | 1418018_at 1418019_at 1418020_s_at 1434547_at 1442212_at 1447392_s_at 1455009_at | 12180 | -0.023 | -0.1725 | No | ||
| 76 | TAL1 | 1449389_at | 12321 | -0.025 | -0.1786 | No | ||
| 77 | MAP3K8 | 1419208_at | 12556 | -0.028 | -0.1889 | No | ||
| 78 | TAZ | 1416207_at 1453785_at | 12561 | -0.028 | -0.1886 | No | ||
| 79 | ERN1 | 1421240_at 1431886_at 1437119_at 1438583_at 1438997_at 1450176_at | 12605 | -0.029 | -0.1901 | No | ||
| 80 | ZBTB5 | 1437484_at | 12628 | -0.029 | -0.1906 | No | ||
| 81 | BFSP1 | 1450571_a_at | 12679 | -0.030 | -0.1924 | No | ||
| 82 | COL16A1 | 1427986_a_at | 12772 | -0.031 | -0.1962 | No | ||
| 83 | C1QL1 | 1422777_at | 12788 | -0.031 | -0.1963 | No | ||
| 84 | SMOC1 | 1448321_at | 12817 | -0.031 | -0.1971 | No | ||
| 85 | GATA4 | 1418863_at 1418864_at 1441364_at | 13034 | -0.035 | -0.2065 | No | ||
| 86 | CXCL2 | 1449984_at | 13143 | -0.036 | -0.2108 | No | ||
| 87 | MSX1 | 1417127_at 1448601_s_at | 13409 | -0.040 | -0.2224 | No | ||
| 88 | CXCL11 | 1419697_at 1419698_at | 13620 | -0.043 | -0.2313 | No | ||
| 89 | PFN1 | 1449018_at | 13688 | -0.044 | -0.2337 | No | ||
| 90 | PRX | 1423292_a_at | 14041 | -0.049 | -0.2490 | No | ||
| 91 | ALG6 | 1439007_at 1459337_at | 14097 | -0.050 | -0.2507 | No | ||
| 92 | BCL6B | 1418421_at | 14247 | -0.053 | -0.2567 | No | ||
| 93 | HOXA11 | 1420414_at | 14272 | -0.053 | -0.2570 | No | ||
| 94 | GRIN2D | 1421393_at | 14774 | -0.061 | -0.2790 | No | ||
| 95 | ASCL3 | 1420780_at | 14794 | -0.062 | -0.2788 | No | ||
| 96 | MAG | 1460219_at | 14803 | -0.062 | -0.2782 | No | ||
| 97 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 14844 | -0.062 | -0.2790 | No | ||
| 98 | ATP1B1 | 1418453_a_at 1423890_x_at 1439036_a_at 1451152_a_at 1455949_at | 15031 | -0.066 | -0.2865 | No | ||
| 99 | AGC1 | 1449827_at | 15150 | -0.067 | -0.2908 | No | ||
| 100 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 15268 | -0.069 | -0.2950 | No | ||
| 101 | UBD | 1419762_at | 15465 | -0.073 | -0.3029 | No | ||
| 102 | WNT10A | 1460657_at | 15513 | -0.074 | -0.3038 | No | ||
| 103 | GREM1 | 1425357_a_at | 15536 | -0.074 | -0.3036 | No | ||
| 104 | VCAM1 | 1415989_at 1436003_at 1448162_at 1451314_a_at | 15580 | -0.075 | -0.3043 | No | ||
| 105 | LIG1 | 1416641_at | 15681 | -0.077 | -0.3077 | No | ||
| 106 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 15698 | -0.077 | -0.3071 | No | ||
| 107 | SESN2 | 1425139_at 1451599_at | 15713 | -0.077 | -0.3065 | No | ||
| 108 | GNG4 | 1417943_at 1417944_at 1447669_s_at | 15800 | -0.079 | -0.3092 | No | ||
| 109 | FLRT1 | 1443143_at | 16029 | -0.083 | -0.3183 | No | ||
| 110 | TLX1 | 1450526_at | 16035 | -0.083 | -0.3172 | No | ||
| 111 | IL1RN | 1423017_a_at 1425663_at 1451798_at | 16076 | -0.084 | -0.3176 | No | ||
| 112 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 16135 | -0.085 | -0.3189 | No | ||
| 113 | NFKB2 | 1425902_a_at 1429128_x_at | 16213 | -0.087 | -0.3210 | No | ||
| 114 | MADCAM1 | 1425253_a_at | 16348 | -0.090 | -0.3257 | No | ||
| 115 | ABHD8 | 1416863_at | 16602 | -0.095 | -0.3358 | No | ||
| 116 | STC2 | 1419503_at 1445186_at 1449484_at | 16926 | -0.103 | -0.3489 | No | ||
| 117 | MSC | 1418417_at | 16998 | -0.105 | -0.3505 | No | ||
| 118 | DAP3 | 1450848_at | 17028 | -0.106 | -0.3500 | No | ||
| 119 | STAT6 | 1421708_a_at 1426353_at | 17159 | -0.109 | -0.3542 | No | ||
| 120 | FKHL18 | 1437820_at | 17185 | -0.110 | -0.3536 | No | ||
| 121 | BAI2 | 1435558_at | 17356 | -0.114 | -0.3595 | No | ||
| 122 | MAML2 | 1446570_at 1447546_s_at | 17423 | -0.116 | -0.3606 | No | ||
| 123 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 17428 | -0.116 | -0.3589 | No | ||
| 124 | CXCL9 | 1418652_at 1456907_at | 17443 | -0.117 | -0.3576 | No | ||
| 125 | BLR1 | 1422003_at | 17551 | -0.120 | -0.3606 | No | ||
| 126 | KLK9 | 1431681_at | 17832 | -0.128 | -0.3713 | No | ||
| 127 | RRAS | 1418448_at | 17876 | -0.129 | -0.3712 | No | ||
| 128 | IL13 | 1420802_at | 17902 | -0.130 | -0.3702 | No | ||
| 129 | CXCL10 | 1418930_at | 17919 | -0.130 | -0.3688 | No | ||
| 130 | RALGDS | 1460634_at | 17946 | -0.131 | -0.3678 | No | ||
| 131 | HIVEP1 | 1422742_at | 18024 | -0.134 | -0.3692 | No | ||
| 132 | RPS6KA4 | 1438243_at 1448498_at | 18177 | -0.138 | -0.3739 | No | ||
| 133 | TNIP1 | 1427689_a_at | 18199 | -0.139 | -0.3726 | No | ||
| 134 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 18294 | -0.142 | -0.3745 | No | ||
| 135 | C1ORF24 | 1422567_at 1454942_at | 18320 | -0.143 | -0.3734 | No | ||
| 136 | EDN2 | 1449161_at | 18341 | -0.143 | -0.3719 | No | ||
| 137 | TNF | 1419607_at | 18434 | -0.146 | -0.3737 | No | ||
| 138 | FOXJ2 | 1420373_at 1420374_at | 18569 | -0.152 | -0.3774 | No | ||
| 139 | UBE2H | 1418631_at 1418632_at 1428791_at 1438971_x_at 1447374_at 1459930_at | 18707 | -0.157 | -0.3811 | No | ||
| 140 | SLAMF8 | 1425294_at | 18856 | -0.164 | -0.3852 | No | ||
| 141 | TCEA2 | 1418107_at 1440791_x_at | 19265 | -0.183 | -0.4010 | No | ||
| 142 | SIN3A | 1419101_at 1419102_at 1419900_at 1449343_s_at | 19293 | -0.185 | -0.3992 | No | ||
| 143 | KCNN3 | 1421632_at 1459308_at | 19418 | -0.191 | -0.4017 | No | ||
| 144 | UBE4B | 1432499_a_at 1444465_at 1448410_at | 19458 | -0.194 | -0.4003 | No | ||
| 145 | SHOX2 | 1420559_a_at 1438042_at | 19595 | -0.203 | -0.4032 | No | ||
| 146 | GRK5 | 1446361_at 1446998_at 1449514_at | 19607 | -0.203 | -0.4004 | No | ||
| 147 | TRAF4 | 1416571_at 1460642_at | 19630 | -0.206 | -0.3980 | No | ||
| 148 | EBI3 | 1449222_at | 19638 | -0.206 | -0.3950 | No | ||
| 149 | USP52 | 1426700_a_at | 20034 | -0.240 | -0.4092 | Yes | ||
| 150 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20040 | -0.240 | -0.4054 | Yes | ||
| 151 | NTN1 | 1422987_at 1454974_at | 20075 | -0.244 | -0.4030 | Yes | ||
| 152 | LRCH1 | 1438032_at 1442502_at 1442757_at 1445396_at 1447746_at 1459150_at | 20164 | -0.255 | -0.4028 | Yes | ||
| 153 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 20223 | -0.261 | -0.4012 | Yes | ||
| 154 | ZBTB9 | 1429125_at | 20433 | -0.287 | -0.4061 | Yes | ||
| 155 | TNFSF7 | 1449926_at | 20459 | -0.290 | -0.4025 | Yes | ||
| 156 | LTB | 1419135_at | 20536 | -0.301 | -0.4010 | Yes | ||
| 157 | JARID2 | 1422698_s_at 1446082_at 1450710_at 1456661_at | 20547 | -0.302 | -0.3965 | Yes | ||
| 158 | ICAM1 | 1424067_at | 20589 | -0.311 | -0.3933 | Yes | ||
| 159 | HCST | 1419119_at | 20631 | -0.317 | -0.3899 | Yes | ||
| 160 | LTA | 1420353_at | 20762 | -0.339 | -0.3903 | Yes | ||
| 161 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 20870 | -0.366 | -0.3892 | Yes | ||
| 162 | UNC84B | 1433832_at 1439238_at | 20884 | -0.370 | -0.3837 | Yes | ||
| 163 | PCSK2 | 1428305_at 1444147_at 1447991_at 1447992_s_at 1448312_at | 20995 | -0.402 | -0.3822 | Yes | ||
| 164 | IRF1 | 1448436_a_at | 20997 | -0.403 | -0.3756 | Yes | ||
| 165 | IL4I1 | 1419192_at | 21061 | -0.427 | -0.3714 | Yes | ||
| 166 | ENO3 | 1417951_at 1449631_at | 21167 | -0.468 | -0.3685 | Yes | ||
| 167 | ZMYND15 | 1438498_at | 21193 | -0.481 | -0.3618 | Yes | ||
| 168 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21244 | -0.512 | -0.3556 | Yes | ||
| 169 | GNAO1 | 1421152_a_at 1448031_at 1460383_at | 21269 | -0.528 | -0.3480 | Yes | ||
| 170 | DCAMKL1 | 1423125_at 1424270_at 1424271_at 1436659_at 1446190_at 1450863_a_at 1451289_at 1451917_a_at | 21296 | -0.547 | -0.3402 | Yes | ||
| 171 | TNFRSF1B | 1418099_at 1448951_at | 21297 | -0.550 | -0.3312 | Yes | ||
| 172 | BDNF | 1422168_a_at 1422169_a_at | 21318 | -0.564 | -0.3228 | Yes | ||
| 173 | SOX10 | 1424985_a_at 1425369_a_at 1451689_a_at | 21378 | -0.616 | -0.3154 | Yes | ||
| 174 | RIMS2 | 1422809_at 1432102_at 1436470_at 1443215_at 1450761_s_at | 21385 | -0.621 | -0.3054 | Yes | ||
| 175 | MSN | 1421814_at 1450379_at | 21392 | -0.627 | -0.2954 | Yes | ||
| 176 | RELB | 1417856_at | 21402 | -0.637 | -0.2853 | Yes | ||
| 177 | CD83 | 1416111_at | 21414 | -0.651 | -0.2751 | Yes | ||
| 178 | IL27RA | 1449508_at | 21476 | -0.706 | -0.2663 | Yes | ||
| 179 | BCL3 | 1418133_at | 21503 | -0.742 | -0.2553 | Yes | ||
| 180 | SLC11A2 | 1417584_at 1426441_at 1441709_at 1447333_at 1452078_a_at | 21575 | -0.827 | -0.2449 | Yes | ||
| 181 | GPHN | 1426462_at 1426463_at 1430038_at 1444869_at 1445188_at 1458242_at | 21690 | -1.083 | -0.2323 | Yes | ||
| 182 | BMF | 1422995_at 1454880_s_at 1458325_x_at | 21713 | -1.175 | -0.2140 | Yes | ||
| 183 | MMP9 | 1416298_at 1448291_at | 21774 | -1.396 | -0.1938 | Yes | ||
| 184 | BIRC3 | 1421392_a_at 1425223_at | 21827 | -1.626 | -0.1694 | Yes | ||
| 185 | JAK3 | 1425750_a_at 1446515_at | 21863 | -1.749 | -0.1422 | Yes | ||
| 186 | CXCL16 | 1418718_at 1449195_s_at | 21890 | -2.020 | -0.1102 | Yes | ||
| 187 | HCFC1 | 1421972_s_at 1433736_at 1445688_at 1446868_at 1450439_at 1457875_at | 21891 | -2.035 | -0.0767 | Yes | ||
| 188 | TNFRSF9 | 1421481_at 1428034_a_at 1460469_at | 21895 | -2.100 | -0.0422 | Yes | ||
| 189 | GNB1 | 1417432_a_at 1425908_at 1454696_at 1459442_at | 21915 | -2.671 | 0.0009 | Yes |

