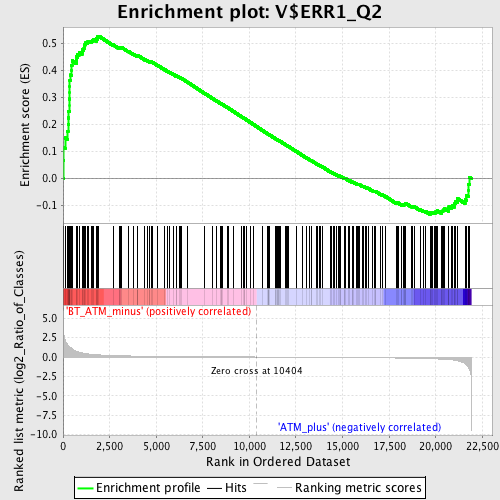
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$ERR1_Q2 |
| Enrichment Score (ES) | 0.52620524 |
| Normalized Enrichment Score (NES) | 1.5315007 |
| Nominal p-value | 0.0013227513 |
| FDR q-value | 0.21024352 |
| FWER p-Value | 0.954 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UHRF2 | 1447217_at 1454920_at 1457070_at | 17 | 3.839 | 0.0664 | Yes | ||
| 2 | RNF139 | 1429425_at 1429426_at | 45 | 2.841 | 0.1148 | Yes | ||
| 3 | MYST3 | 1436315_at 1437487_at 1446143_at 1446405_at | 109 | 2.189 | 0.1502 | Yes | ||
| 4 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 222 | 1.655 | 0.1740 | Yes | ||
| 5 | SRPK2 | 1417134_at 1417135_at 1417136_s_at 1431372_at 1435746_at 1440080_at 1442241_at 1442980_at 1447755_at 1448603_at | 268 | 1.526 | 0.1986 | Yes | ||
| 6 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 284 | 1.486 | 0.2239 | Yes | ||
| 7 | RYR3 | 1427427_at 1452533_at | 301 | 1.453 | 0.2486 | Yes | ||
| 8 | ABCC5 | 1418042_a_at 1418043_at 1427565_a_at 1435683_a_at 1435684_at 1435685_x_at 1438056_x_at 1447384_at 1459782_x_at | 340 | 1.350 | 0.2704 | Yes | ||
| 9 | NR2C2 | 1425014_at 1439696_at 1445613_at 1446955_at 1451569_at 1454167_at 1454851_at | 342 | 1.348 | 0.2939 | Yes | ||
| 10 | TBC1D15 | 1416060_at 1416061_at 1416062_at | 353 | 1.322 | 0.3166 | Yes | ||
| 11 | ATP6V1A | 1422508_at 1445916_at 1450634_at | 357 | 1.302 | 0.3392 | Yes | ||
| 12 | VSNL1 | 1420955_at 1450055_at | 362 | 1.289 | 0.3616 | Yes | ||
| 13 | MSI2 | 1421230_a_at 1421231_at 1435520_at 1435521_at 1436818_a_at 1441185_at 1441373_at 1443008_at 1443530_at 1446022_at 1458136_at 1458542_at | 382 | 1.253 | 0.3826 | Yes | ||
| 14 | SEC61A1 | 1416189_a_at 1416190_a_at 1416191_at 1434986_a_at 1448242_at 1455987_at | 450 | 1.121 | 0.3992 | Yes | ||
| 15 | USP8 | 1422883_at 1443420_at 1446807_at 1447386_at | 457 | 1.110 | 0.4183 | Yes | ||
| 16 | JARID1A | 1437105_at 1437106_at 1441318_at 1444842_at 1445541_at 1452360_a_at | 482 | 1.039 | 0.4354 | Yes | ||
| 17 | PHB | 1435144_at 1442233_at 1448563_at | 706 | 0.739 | 0.4380 | Yes | ||
| 18 | ANKRD28 | 1434630_at 1454801_at | 728 | 0.719 | 0.4496 | Yes | ||
| 19 | AFG3L2 | 1427206_at 1427207_s_at 1454003_at | 766 | 0.683 | 0.4599 | Yes | ||
| 20 | PPM1E | 1434990_at 1445741_at | 855 | 0.624 | 0.4667 | Yes | ||
| 21 | SLC31A2 | 1416654_at 1453721_a_at 1459399_at | 1018 | 0.528 | 0.4685 | Yes | ||
| 22 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 1047 | 0.513 | 0.4762 | Yes | ||
| 23 | SATB1 | 1416007_at 1416008_at 1439203_at 1439449_at 1443284_at 1456902_at | 1117 | 0.485 | 0.4815 | Yes | ||
| 24 | LDB3 | 1416752_at 1433783_at 1451999_at | 1139 | 0.474 | 0.4888 | Yes | ||
| 25 | VCL | 1416156_at 1416157_at 1445256_at | 1169 | 0.463 | 0.4956 | Yes | ||
| 26 | ZDHHC21 | 1429488_at | 1219 | 0.446 | 0.5012 | Yes | ||
| 27 | PANK1 | 1418715_at 1429813_at 1429814_at 1431028_a_at 1457110_at | 1300 | 0.411 | 0.5047 | Yes | ||
| 28 | HTATSF1 | 1431058_at 1431059_x_at 1454760_at | 1383 | 0.384 | 0.5076 | Yes | ||
| 29 | RAP2C | 1428119_a_at 1437016_x_at 1460430_at | 1511 | 0.351 | 0.5079 | Yes | ||
| 30 | SEMA7A | 1422040_at 1459903_at | 1583 | 0.335 | 0.5105 | Yes | ||
| 31 | YTHDF2 | 1451317_at 1460357_at | 1620 | 0.328 | 0.5146 | Yes | ||
| 32 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 1777 | 0.299 | 0.5127 | Yes | ||
| 33 | SUCLA2 | 1430402_at 1452206_at | 1804 | 0.293 | 0.5166 | Yes | ||
| 34 | PHF6 | 1430415_at 1453761_at 1454625_at | 1838 | 0.287 | 0.5201 | Yes | ||
| 35 | HSPD1 | 1426351_at | 1860 | 0.283 | 0.5241 | Yes | ||
| 36 | ATP5G1 | 1416020_a_at 1444874_at | 1918 | 0.273 | 0.5262 | Yes | ||
| 37 | SNTG1 | 1432287_a_at 1440598_at | 2681 | 0.184 | 0.4944 | No | ||
| 38 | PRKG2 | 1421354_at 1435162_at 1435460_at | 3007 | 0.161 | 0.4823 | No | ||
| 39 | SLC37A2 | 1452492_a_at | 3013 | 0.160 | 0.4848 | No | ||
| 40 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 3064 | 0.157 | 0.4853 | No | ||
| 41 | LHX2 | 1418317_at | 3147 | 0.153 | 0.4842 | No | ||
| 42 | KIF5A | 1434670_at | 3511 | 0.135 | 0.4699 | No | ||
| 43 | CSNK2A1 | 1419034_at 1419035_s_at 1419036_at 1419037_at 1419038_a_at 1453427_at | 3787 | 0.124 | 0.4594 | No | ||
| 44 | APP | 1420621_a_at 1427442_a_at 1438373_at 1438374_x_at 1444705_at 1447347_at 1458525_at | 3980 | 0.116 | 0.4526 | No | ||
| 45 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 4000 | 0.115 | 0.4537 | No | ||
| 46 | GRIK3 | 1427709_at | 4006 | 0.115 | 0.4555 | No | ||
| 47 | PSIP1 | 1417166_at 1442148_at 1460403_at | 4395 | 0.102 | 0.4395 | No | ||
| 48 | PCDH21 | 1418304_at | 4508 | 0.100 | 0.4360 | No | ||
| 49 | SCNN1A | 1425088_at | 4653 | 0.096 | 0.4311 | No | ||
| 50 | HDAC11 | 1451229_at 1454803_a_at | 4660 | 0.096 | 0.4325 | No | ||
| 51 | KCNG4 | 1428536_at | 4728 | 0.094 | 0.4311 | No | ||
| 52 | PIM1 | 1423006_at 1435458_at | 4778 | 0.092 | 0.4304 | No | ||
| 53 | SART3 | 1417547_at 1417548_at | 5077 | 0.085 | 0.4182 | No | ||
| 54 | RNF2 | 1424873_at 1448041_at 1448042_s_at 1448043_x_at 1451519_at | 5455 | 0.077 | 0.4022 | No | ||
| 55 | RHCG | 1417362_at | 5605 | 0.074 | 0.3967 | No | ||
| 56 | RASAL1 | 1417858_at | 5726 | 0.071 | 0.3924 | No | ||
| 57 | HECTD2 | 1433944_at 1438596_at 1446224_at 1446430_at | 5942 | 0.067 | 0.3837 | No | ||
| 58 | SPATS1 | 1432238_at 1443710_s_at | 6064 | 0.065 | 0.3793 | No | ||
| 59 | LIMK1 | 1417627_a_at 1425836_a_at 1456234_at 1458989_at | 6074 | 0.065 | 0.3800 | No | ||
| 60 | HSPE1 | 1450668_s_at | 6259 | 0.061 | 0.3726 | No | ||
| 61 | KIRREL3 | 1431402_at 1441357_at 1452728_at | 6294 | 0.060 | 0.3721 | No | ||
| 62 | RAB3A | 1422589_at 1459980_x_at | 6345 | 0.059 | 0.3708 | No | ||
| 63 | CHD2 | 1444246_at 1445843_at | 6687 | 0.054 | 0.3561 | No | ||
| 64 | FZD9 | 1427529_at | 7607 | 0.039 | 0.3145 | No | ||
| 65 | NAV1 | 1436907_at 1443629_at 1446497_at | 8013 | 0.033 | 0.2964 | No | ||
| 66 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 8254 | 0.029 | 0.2859 | No | ||
| 67 | ITGA3 | 1421997_s_at 1455158_at 1460305_at | 8439 | 0.026 | 0.2779 | No | ||
| 68 | KCNK1 | 1448690_at 1455896_a_at | 8483 | 0.026 | 0.2764 | No | ||
| 69 | SDHB | 1418005_at | 8506 | 0.026 | 0.2758 | No | ||
| 70 | ADCY1 | 1445112_at 1445359_at 1456487_at 1457251_x_at 1457329_at | 8534 | 0.025 | 0.2750 | No | ||
| 71 | VDAC1 | 1415998_at 1436992_x_at 1437192_x_at 1437452_x_at 1437947_x_at | 8814 | 0.021 | 0.2626 | No | ||
| 72 | UQCRC1 | 1428782_a_at | 8888 | 0.020 | 0.2596 | No | ||
| 73 | NEF3 | 1422520_at | 9170 | 0.017 | 0.2469 | No | ||
| 74 | STRN | 1422036_at 1455022_at 1455156_at | 9599 | 0.011 | 0.2274 | No | ||
| 75 | SSR4 | 1427096_s_at 1435709_at 1448524_s_at | 9680 | 0.010 | 0.2239 | No | ||
| 76 | RIMS4 | 1436564_at 1456926_at | 9713 | 0.009 | 0.2226 | No | ||
| 77 | ACTR1A | 1415873_a_at 1458696_at 1459855_x_at | 9715 | 0.009 | 0.2227 | No | ||
| 78 | WBSCR1 | 1416941_s_at 1436637_at 1438554_x_at 1442942_at 1447070_at 1451521_x_at | 9859 | 0.007 | 0.2163 | No | ||
| 79 | EPN3 | 1424960_at | 10061 | 0.004 | 0.2071 | No | ||
| 80 | HS6ST2 | 1420938_at 1420939_at 1446257_at 1450047_at | 10235 | 0.002 | 0.1992 | No | ||
| 81 | SLC30A3 | 1460654_at | 10719 | -0.004 | 0.1771 | No | ||
| 82 | SHB | 1434153_at | 10994 | -0.008 | 0.1646 | No | ||
| 83 | PHF5A | 1424170_at | 11027 | -0.008 | 0.1633 | No | ||
| 84 | TCF2 | 1421224_a_at 1441484_at 1451687_a_at | 11043 | -0.008 | 0.1627 | No | ||
| 85 | RAPGEF5 | 1445255_at 1455137_at 1455840_at | 11048 | -0.009 | 0.1627 | No | ||
| 86 | ASCC1 | 1438539_at 1442874_at 1460703_at | 11084 | -0.009 | 0.1612 | No | ||
| 87 | MB | 1422420_at 1451203_at | 11378 | -0.013 | 0.1480 | No | ||
| 88 | SCN5A | 1422194_at 1458813_at | 11415 | -0.013 | 0.1466 | No | ||
| 89 | SORCS1 | 1421509_at 1425864_a_at 1436662_at | 11467 | -0.014 | 0.1445 | No | ||
| 90 | TGM6 | 1437702_at | 11508 | -0.014 | 0.1429 | No | ||
| 91 | NXPH4 | 1429753_at | 11532 | -0.015 | 0.1421 | No | ||
| 92 | PVALB | 1417653_at | 11543 | -0.015 | 0.1419 | No | ||
| 93 | SAG | 1419025_at | 11559 | -0.015 | 0.1414 | No | ||
| 94 | MMP24 | 1417678_at 1441456_at 1442921_at 1448806_at 1456376_at | 11609 | -0.016 | 0.1395 | No | ||
| 95 | ABAT | 1433855_at | 11697 | -0.017 | 0.1358 | No | ||
| 96 | DIRAS1 | 1425748_at 1451820_at 1455526_at | 11917 | -0.019 | 0.1260 | No | ||
| 97 | VAMP1 | 1421862_a_at 1421863_at | 11978 | -0.020 | 0.1236 | No | ||
| 98 | FGF9 | 1420795_at 1438718_at 1445258_at | 12025 | -0.021 | 0.1219 | No | ||
| 99 | PLCB3 | 1448661_at | 12112 | -0.022 | 0.1183 | No | ||
| 100 | CDH16 | 1448906_at | 12534 | -0.028 | 0.0994 | No | ||
| 101 | RNF128 | 1418318_at 1449036_at | 12874 | -0.032 | 0.0844 | No | ||
| 102 | FBXL19 | 1435922_at | 13086 | -0.035 | 0.0753 | No | ||
| 103 | OBSCN | 1443632_at | 13236 | -0.037 | 0.0691 | No | ||
| 104 | ADAM9 | 1416094_at | 13337 | -0.039 | 0.0652 | No | ||
| 105 | TIMM8B | 1431665_a_at | 13339 | -0.039 | 0.0658 | No | ||
| 106 | SOD3 | 1417633_at 1417634_at | 13581 | -0.043 | 0.0555 | No | ||
| 107 | GEMIN7 | 1435764_a_at | 13679 | -0.044 | 0.0518 | No | ||
| 108 | ELAVL3 | 1452524_a_at | 13770 | -0.045 | 0.0485 | No | ||
| 109 | WDFY3 | 1427456_at 1428517_at 1445718_at 1446109_at 1447433_at 1458952_at 1459287_at 1459414_at | 13804 | -0.046 | 0.0477 | No | ||
| 110 | RPH3A | 1434635_at 1449961_at | 13926 | -0.048 | 0.0430 | No | ||
| 111 | CLPTM1 | 1416883_at | 14359 | -0.055 | 0.0241 | No | ||
| 112 | NTF3 | 1434802_s_at 1450803_at | 14438 | -0.056 | 0.0215 | No | ||
| 113 | MYL3 | 1427768_s_at 1427769_x_at 1428266_at | 14496 | -0.057 | 0.0199 | No | ||
| 114 | STAC2 | 1424581_at | 14581 | -0.058 | 0.0170 | No | ||
| 115 | SPAG9 | 1430556_at 1433648_at 1437676_at | 14688 | -0.060 | 0.0132 | No | ||
| 116 | UQCRC2 | 1428631_a_at 1435757_a_at | 14792 | -0.062 | 0.0096 | No | ||
| 117 | FBS1 | 1425276_at 1451645_at | 14819 | -0.062 | 0.0094 | No | ||
| 118 | PPP2R5D | 1450560_a_at | 14894 | -0.063 | 0.0071 | No | ||
| 119 | ESRRA | 1442864_at 1460652_at | 14903 | -0.063 | 0.0079 | No | ||
| 120 | UNC5B | 1435110_at 1453269_at | 15104 | -0.067 | -0.0001 | No | ||
| 121 | KCNH2 | 1449544_a_at | 15128 | -0.067 | -0.0000 | No | ||
| 122 | IDH3G | 1416788_a_at 1416789_at | 15141 | -0.067 | 0.0006 | No | ||
| 123 | KCNN1 | 1419617_at 1449536_at | 15318 | -0.070 | -0.0063 | No | ||
| 124 | ACO2 | 1436934_s_at 1442102_at 1451002_at | 15399 | -0.072 | -0.0087 | No | ||
| 125 | NOG | 1422300_at | 15517 | -0.074 | -0.0128 | No | ||
| 126 | PSCD2 | 1450103_a_at | 15556 | -0.074 | -0.0132 | No | ||
| 127 | MAP1A | 1460566_at | 15591 | -0.075 | -0.0135 | No | ||
| 128 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 15770 | -0.078 | -0.0203 | No | ||
| 129 | CXXC5 | 1431469_a_at 1448960_at | 15807 | -0.079 | -0.0206 | No | ||
| 130 | TCF7 | 1433471_at 1450461_at | 15848 | -0.080 | -0.0210 | No | ||
| 131 | KIF3B | 1420987_at 1450073_at 1450074_at | 15892 | -0.081 | -0.0216 | No | ||
| 132 | THRA | 1426997_at 1443952_at 1454675_at | 16064 | -0.084 | -0.0280 | No | ||
| 133 | SCNN1G | 1419079_at | 16122 | -0.085 | -0.0291 | No | ||
| 134 | MRPL48 | 1428302_at 1437997_x_at | 16251 | -0.087 | -0.0335 | No | ||
| 135 | DLC1 | 1436173_at 1450206_at 1458220_at 1460602_at | 16311 | -0.089 | -0.0347 | No | ||
| 136 | CNTN2 | 1435165_at 1435166_at 1450523_at 1456962_at | 16382 | -0.090 | -0.0363 | No | ||
| 137 | LMO3 | 1455754_at | 16590 | -0.095 | -0.0442 | No | ||
| 138 | SLC25A5 | 1423772_x_at 1434801_x_at 1436874_x_at 1445944_at | 16700 | -0.097 | -0.0475 | No | ||
| 139 | LRFN4 | 1454700_at | 16755 | -0.099 | -0.0482 | No | ||
| 140 | ZRANB1 | 1415712_at 1427200_at 1436228_at | 16774 | -0.099 | -0.0473 | No | ||
| 141 | HCRTR1 | 1436295_at | 17062 | -0.107 | -0.0586 | No | ||
| 142 | TEF | 1424175_at 1438033_at 1450184_s_at | 17146 | -0.109 | -0.0606 | No | ||
| 143 | FGL1 | 1424599_at | 17322 | -0.113 | -0.0666 | No | ||
| 144 | TIMM9 | 1428010_at 1449886_a_at | 17894 | -0.130 | -0.0906 | No | ||
| 145 | RAB35 | 1433922_at | 17938 | -0.131 | -0.0903 | No | ||
| 146 | PDCD8 | 1418127_a_at | 17989 | -0.132 | -0.0903 | No | ||
| 147 | FRAG1 | 1424614_at 1424615_at 1424616_s_at | 18175 | -0.138 | -0.0964 | No | ||
| 148 | VDAC2 | 1415990_at | 18192 | -0.139 | -0.0947 | No | ||
| 149 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 18294 | -0.142 | -0.0969 | No | ||
| 150 | ATP6AP2 | 1423662_at 1437688_x_at 1439456_x_at | 18323 | -0.143 | -0.0956 | No | ||
| 151 | UCHL1 | 1448260_at | 18357 | -0.144 | -0.0946 | No | ||
| 152 | AFP | 1416645_a_at 1416646_at 1436879_x_at 1436880_at 1436881_x_at | 18394 | -0.145 | -0.0938 | No | ||
| 153 | TFEB | 1422566_at | 18409 | -0.146 | -0.0919 | No | ||
| 154 | SNAG1 | 1416359_at 1416360_at | 18708 | -0.157 | -0.1028 | No | ||
| 155 | MGST3 | 1448300_at 1450563_at | 18786 | -0.160 | -0.1036 | No | ||
| 156 | RNF44 | 1423532_at | 18863 | -0.164 | -0.1042 | No | ||
| 157 | MNT | 1418192_at | 19190 | -0.179 | -0.1160 | No | ||
| 158 | BCKDHB | 1427153_at | 19358 | -0.188 | -0.1204 | No | ||
| 159 | IBRDC3 | 1432478_a_at 1435226_at | 19446 | -0.194 | -0.1210 | No | ||
| 160 | SOCS4 | 1421273_at 1421274_at 1421275_s_at 1455142_at | 19709 | -0.212 | -0.1294 | No | ||
| 161 | ATP5J2 | 1416269_at 1435395_s_at | 19724 | -0.213 | -0.1263 | No | ||
| 162 | GJB2 | 1423271_at | 19766 | -0.215 | -0.1244 | No | ||
| 163 | VAMP2 | 1420833_at 1420834_at | 19854 | -0.222 | -0.1246 | No | ||
| 164 | SMPD1 | 1447874_x_at 1448621_a_at | 19961 | -0.232 | -0.1254 | No | ||
| 165 | RGS4 | 1416286_at 1416287_at 1448285_at | 20016 | -0.238 | -0.1237 | No | ||
| 166 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20040 | -0.240 | -0.1206 | No | ||
| 167 | SMARCC2 | 1428382_at | 20095 | -0.246 | -0.1187 | No | ||
| 168 | NDRG2 | 1448154_at | 20335 | -0.274 | -0.1249 | No | ||
| 169 | SDHD | 1428235_at 1437489_x_at | 20344 | -0.275 | -0.1205 | No | ||
| 170 | HOXA5 | 1443803_x_at 1448926_at | 20398 | -0.283 | -0.1180 | No | ||
| 171 | RFX5 | 1423103_at | 20449 | -0.290 | -0.1152 | No | ||
| 172 | GOT1 | 1450970_at | 20499 | -0.295 | -0.1123 | No | ||
| 173 | RASGRP2 | 1417804_at 1438932_at 1438933_x_at 1442264_at 1449613_at | 20690 | -0.327 | -0.1153 | No | ||
| 174 | FBXL17 | 1430241_at 1452860_at | 20703 | -0.329 | -0.1101 | No | ||
| 175 | ID3 | 1416630_at | 20719 | -0.332 | -0.1050 | No | ||
| 176 | JARID1C | 1426497_at 1426498_at 1441449_at 1441450_s_at 1444157_a_at 1444158_at 1457930_at | 20853 | -0.361 | -0.1048 | No | ||
| 177 | VASP | 1451097_at | 20921 | -0.381 | -0.1012 | No | ||
| 178 | CACNB2 | 1441521_at 1444693_at 1446632_at 1452476_at 1456401_at | 21018 | -0.412 | -0.0984 | No | ||
| 179 | BZW2 | 1423456_at 1443227_at | 21036 | -0.420 | -0.0919 | No | ||
| 180 | SLC38A3 | 1418706_at 1446013_at | 21073 | -0.431 | -0.0860 | No | ||
| 181 | ENO3 | 1417951_at 1449631_at | 21167 | -0.468 | -0.0821 | No | ||
| 182 | XYLT2 | 1452515_a_at | 21170 | -0.469 | -0.0739 | No | ||
| 183 | NR4A1 | 1416505_at | 21587 | -0.852 | -0.0782 | No | ||
| 184 | ANGPT1 | 1421441_at 1438936_s_at 1438937_x_at 1439066_at 1446656_at 1450717_at 1457381_at | 21667 | -1.017 | -0.0640 | No | ||
| 185 | TMEM28 | 1435138_at 1443271_at 1445869_at 1446912_at 1459483_at | 21761 | -1.355 | -0.0446 | No | ||
| 186 | PCDH7 | 1427592_at 1437442_at 1449249_at 1456214_at | 21768 | -1.369 | -0.0209 | No | ||
| 187 | PHF15 | 1430081_at 1455345_at | 21828 | -1.631 | 0.0049 | No |

