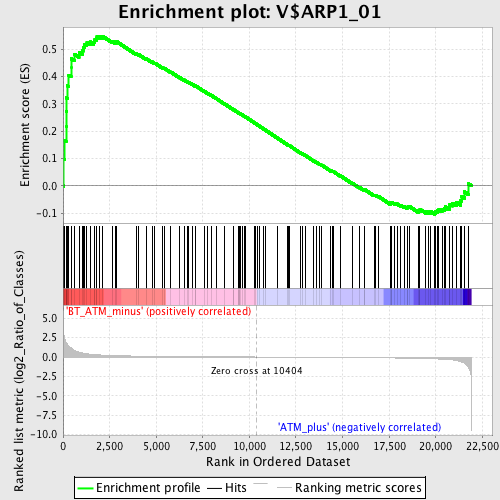
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$ARP1_01 |
| Enrichment Score (ES) | 0.5473651 |
| Normalized Enrichment Score (NES) | 1.5106091 |
| Nominal p-value | 0.011204482 |
| FDR q-value | 0.21360317 |
| FWER p-Value | 0.982 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FBXL20 | 1442464_at 1445575_at 1452826_s_at 1456378_s_at | 26 | 3.426 | 0.0978 | Yes | ||
| 2 | LTBP3 | 1418049_at 1437833_at 1438312_s_at 1456189_x_at | 82 | 2.475 | 0.1668 | Yes | ||
| 3 | MED8 | 1431423_a_at 1442417_at | 161 | 1.889 | 0.2179 | Yes | ||
| 4 | ENSA | 1423330_at 1428620_at | 168 | 1.850 | 0.2711 | Yes | ||
| 5 | PIK3CG | 1422707_at 1422708_at | 187 | 1.787 | 0.3219 | Yes | ||
| 6 | CDC37L1 | 1417740_at 1430205_a_at 1433574_at 1440886_at 1447753_at | 242 | 1.605 | 0.3658 | Yes | ||
| 7 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 307 | 1.436 | 0.4044 | Yes | ||
| 8 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 429 | 1.159 | 0.4323 | Yes | ||
| 9 | PITPNC1 | 1428025_s_at 1428878_a_at 1428879_at 1431074_a_at 1435066_at 1441574_at 1452939_a_at 1452940_x_at 1453750_x_at 1455204_at | 433 | 1.147 | 0.4653 | Yes | ||
| 10 | RNF34 | 1415791_at 1458064_at | 611 | 0.848 | 0.4817 | Yes | ||
| 11 | PPM1E | 1434990_at 1445741_at | 855 | 0.624 | 0.4886 | Yes | ||
| 12 | HRBL | 1425362_at 1437642_at 1441347_at | 1031 | 0.521 | 0.4957 | Yes | ||
| 13 | NXPH1 | 1436718_at 1438388_at 1459921_at | 1092 | 0.494 | 0.5072 | Yes | ||
| 14 | APOM | 1419095_a_at 1419096_at | 1157 | 0.467 | 0.5177 | Yes | ||
| 15 | NFE2 | 1452001_at | 1280 | 0.421 | 0.5243 | Yes | ||
| 16 | NR5A2 | 1420410_at 1449706_s_at 1449707_at | 1465 | 0.360 | 0.5263 | Yes | ||
| 17 | CTLA4 | 1419334_at | 1658 | 0.322 | 0.5268 | Yes | ||
| 18 | HAPLN1 | 1421633_a_at 1426294_at 1426295_at 1438020_at 1445735_at 1458109_at | 1707 | 0.311 | 0.5336 | Yes | ||
| 19 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 1777 | 0.299 | 0.5390 | Yes | ||
| 20 | CDK5R1 | 1421123_at 1421124_at 1433450_at 1433451_at | 1817 | 0.291 | 0.5457 | Yes | ||
| 21 | JPH1 | 1421520_at 1438428_at | 1950 | 0.268 | 0.5474 | Yes | ||
| 22 | EMP1 | 1416529_at 1459171_at | 2117 | 0.245 | 0.5469 | No | ||
| 23 | TRERF1 | 1444654_at | 2651 | 0.186 | 0.5278 | No | ||
| 24 | POU3F4 | 1422164_at 1422165_at | 2811 | 0.174 | 0.5256 | No | ||
| 25 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 2878 | 0.169 | 0.5274 | No | ||
| 26 | CFL2 | 1418066_at 1418067_at 1431432_at | 3926 | 0.118 | 0.4829 | No | ||
| 27 | GPR3 | 1460275_at | 4063 | 0.113 | 0.4799 | No | ||
| 28 | MYOCD | 1425808_a_at 1425978_at | 4463 | 0.101 | 0.4645 | No | ||
| 29 | CASK | 1422518_at 1422519_at 1427692_a_at 1445152_at | 4774 | 0.092 | 0.4530 | No | ||
| 30 | CTGF | 1416953_at | 4924 | 0.089 | 0.4487 | No | ||
| 31 | ATBF1 | 1420649_at 1420650_at 1429725_at 1445941_at 1446094_at 1449947_s_at 1453267_at 1460006_at | 5325 | 0.080 | 0.4327 | No | ||
| 32 | ZFX | 1419565_a_at 1425972_a_at 1428046_a_at 1449512_a_at | 5433 | 0.077 | 0.4301 | No | ||
| 33 | RIN1 | 1424507_at 1437460_x_at | 5778 | 0.070 | 0.4163 | No | ||
| 34 | MYL1 | 1452651_a_at | 6261 | 0.061 | 0.3960 | No | ||
| 35 | OMD | 1418745_at | 6492 | 0.057 | 0.3871 | No | ||
| 36 | CHD2 | 1444246_at 1445843_at | 6687 | 0.054 | 0.3798 | No | ||
| 37 | BTBD5 | 1428721_at 1453987_at | 6744 | 0.053 | 0.3787 | No | ||
| 38 | SLC7A8 | 1417929_at | 6946 | 0.049 | 0.3709 | No | ||
| 39 | ARR3 | 1425232_x_at 1441144_at 1450329_a_at | 7088 | 0.047 | 0.3658 | No | ||
| 40 | AQP7 | 1418848_at 1418849_x_at | 7113 | 0.046 | 0.3661 | No | ||
| 41 | SPARC | 1416589_at 1448392_at 1458204_at | 7572 | 0.039 | 0.3462 | No | ||
| 42 | TNRC4 | 1452851_at | 7757 | 0.037 | 0.3388 | No | ||
| 43 | HOXD4 | 1450209_at | 7968 | 0.033 | 0.3302 | No | ||
| 44 | MNAT1 | 1432177_a_at | 8233 | 0.029 | 0.3189 | No | ||
| 45 | IRAK1 | 1448668_a_at 1460649_at | 8664 | 0.023 | 0.2999 | No | ||
| 46 | HOXA1 | 1420565_at | 9131 | 0.017 | 0.2790 | No | ||
| 47 | B3GALT2 | 1423084_at 1437433_at 1437644_at | 9172 | 0.017 | 0.2777 | No | ||
| 48 | HPCA | 1425833_a_at 1450930_at | 9424 | 0.013 | 0.2665 | No | ||
| 49 | HTR1B | 1422288_at | 9435 | 0.013 | 0.2665 | No | ||
| 50 | RASIP1 | 1428016_a_at | 9455 | 0.013 | 0.2660 | No | ||
| 51 | STAR | 1418728_at 1418729_at 1449201_at | 9492 | 0.012 | 0.2647 | No | ||
| 52 | CLDN15 | 1418920_at | 9535 | 0.012 | 0.2631 | No | ||
| 53 | BMP5 | 1421282_at 1421283_at 1444470_x_at 1455851_at | 9630 | 0.010 | 0.2591 | No | ||
| 54 | FGF20 | 1421677_at | 9766 | 0.008 | 0.2531 | No | ||
| 55 | ESRRG | 1421747_at 1455267_at 1457896_at | 9793 | 0.008 | 0.2522 | No | ||
| 56 | IL21 | 1450334_at | 10284 | 0.002 | 0.2298 | No | ||
| 57 | CDX1 | 1449582_at | 10356 | 0.001 | 0.2265 | No | ||
| 58 | SKIL | 1422054_a_at 1452214_at | 10423 | -0.000 | 0.2235 | No | ||
| 59 | PDZRN4 | 1456512_at | 10551 | -0.002 | 0.2177 | No | ||
| 60 | ELF4 | 1421337_at 1445778_at | 10766 | -0.005 | 0.2081 | No | ||
| 61 | CYP2E1 | 1415994_at | 10857 | -0.006 | 0.2041 | No | ||
| 62 | SERPINC1 | 1417909_at | 11534 | -0.015 | 0.1735 | No | ||
| 63 | DLX5 | 1449863_a_at | 12069 | -0.021 | 0.1497 | No | ||
| 64 | OTX2 | 1425926_a_at | 12127 | -0.022 | 0.1477 | No | ||
| 65 | CACNA1G | 1423365_at | 12159 | -0.023 | 0.1470 | No | ||
| 66 | COL16A1 | 1427986_a_at | 12772 | -0.031 | 0.1198 | No | ||
| 67 | ARHGEF15 | 1440358_at 1455522_at 1457172_at | 12852 | -0.032 | 0.1171 | No | ||
| 68 | DNAJC5B | 1418725_at | 13007 | -0.034 | 0.1110 | No | ||
| 69 | NR6A1 | 1421515_at 1421516_at 1428825_at 1428826_at 1445872_at | 13465 | -0.041 | 0.0913 | No | ||
| 70 | ASB12 | 1420346_at | 13590 | -0.043 | 0.0868 | No | ||
| 71 | PROP1 | 1450361_at | 13786 | -0.046 | 0.0792 | No | ||
| 72 | KIF13A | 1439532_s_at 1440578_at 1442252_at 1446026_at 1447853_x_at 1451890_at 1455746_at | 13898 | -0.047 | 0.0755 | No | ||
| 73 | RALY | 1418573_a_at | 14380 | -0.055 | 0.0550 | No | ||
| 74 | FCHSD1 | 1427155_at | 14449 | -0.056 | 0.0535 | No | ||
| 75 | NFIX | 1423493_a_at 1436363_a_at 1436364_x_at 1451443_at 1459909_at | 14504 | -0.057 | 0.0527 | No | ||
| 76 | E2F4 | 1451480_at | 14891 | -0.063 | 0.0369 | No | ||
| 77 | TGFB3 | 1417455_at | 15532 | -0.074 | 0.0097 | No | ||
| 78 | SLITRK1 | 1428089_at | 15916 | -0.081 | -0.0055 | No | ||
| 79 | SLITRK6 | 1437231_at | 16167 | -0.086 | -0.0145 | No | ||
| 80 | PKP3 | 1418831_at 1418832_at | 16189 | -0.086 | -0.0130 | No | ||
| 81 | DLX1 | 1449470_at | 16705 | -0.098 | -0.0338 | No | ||
| 82 | PDGFB | 1450413_at 1450414_at | 16775 | -0.099 | -0.0341 | No | ||
| 83 | SLC12A5 | 1425337_at 1445933_at 1451674_at | 16922 | -0.103 | -0.0378 | No | ||
| 84 | CCDC6 | 1428311_at 1459159_a_at | 17591 | -0.121 | -0.0649 | No | ||
| 85 | MRPS18B | 1451164_a_at | 17617 | -0.121 | -0.0625 | No | ||
| 86 | C1QTNF7 | 1429030_at 1460163_at | 17641 | -0.122 | -0.0601 | No | ||
| 87 | MAB21L2 | 1418934_at | 17805 | -0.127 | -0.0639 | No | ||
| 88 | FOXP2 | 1422014_at 1438231_at 1438232_at 1440108_at 1441365_at 1458191_at | 17936 | -0.131 | -0.0660 | No | ||
| 89 | CPNE1 | 1433439_at | 18111 | -0.136 | -0.0701 | No | ||
| 90 | GPR124 | 1418379_s_at | 18316 | -0.142 | -0.0753 | No | ||
| 91 | PMF1 | 1416707_a_at 1438173_x_at | 18473 | -0.148 | -0.0782 | No | ||
| 92 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 18519 | -0.150 | -0.0759 | No | ||
| 93 | FCHSD2 | 1434260_at 1456949_at | 18623 | -0.154 | -0.0762 | No | ||
| 94 | PROSAPIP1 | 1441963_at | 19095 | -0.174 | -0.0928 | No | ||
| 95 | DBP | 1418174_at 1438211_s_at | 19121 | -0.175 | -0.0888 | No | ||
| 96 | NDST2 | 1417931_at | 19161 | -0.177 | -0.0855 | No | ||
| 97 | TGIF2 | 1431115_at 1434400_at | 19479 | -0.196 | -0.0944 | No | ||
| 98 | TRAF4 | 1416571_at 1460642_at | 19630 | -0.206 | -0.0953 | No | ||
| 99 | POLD4 | 1427885_at | 19721 | -0.213 | -0.0933 | No | ||
| 100 | BAHD1 | 1433703_s_at | 19968 | -0.233 | -0.0978 | No | ||
| 101 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 20012 | -0.237 | -0.0929 | No | ||
| 102 | SMARCC2 | 1428382_at | 20095 | -0.246 | -0.0896 | No | ||
| 103 | AP1B1 | 1416279_at | 20179 | -0.257 | -0.0860 | No | ||
| 104 | RUNX3 | 1421467_at 1440275_at | 20351 | -0.276 | -0.0858 | No | ||
| 105 | RBBP6 | 1425114_at 1425115_at 1425421_at 1426487_a_at 1453611_at | 20476 | -0.292 | -0.0831 | No | ||
| 106 | GATA1 | 1449232_at | 20523 | -0.299 | -0.0765 | No | ||
| 107 | HSPB8 | 1417013_at 1417014_at 1456434_x_at | 20746 | -0.337 | -0.0770 | No | ||
| 108 | SIPA1 | 1416206_at | 20767 | -0.340 | -0.0680 | No | ||
| 109 | PLK3 | 1434496_at | 20890 | -0.372 | -0.0629 | No | ||
| 110 | ZHX2 | 1455210_at | 21117 | -0.450 | -0.0602 | No | ||
| 111 | RRAGD | 1431164_at 1434909_at 1440555_at | 21357 | -0.593 | -0.0540 | No | ||
| 112 | TREML2 | 1419837_at 1440298_at 1440938_at 1444718_at | 21393 | -0.627 | -0.0375 | No | ||
| 113 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 21554 | -0.797 | -0.0218 | No | ||
| 114 | TMEM28 | 1435138_at 1443271_at 1445869_at 1446912_at 1459483_at | 21761 | -1.355 | 0.0079 | No |

