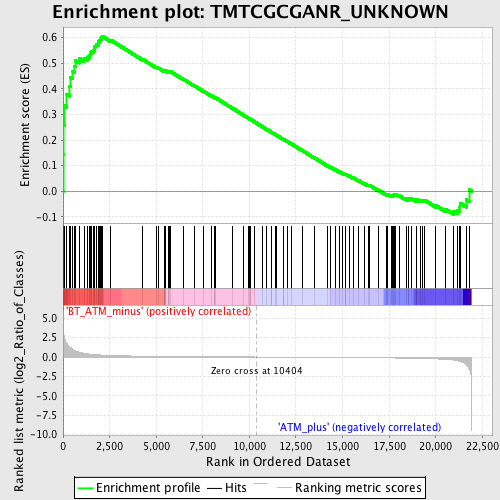
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | TMTCGCGANR_UNKNOWN |
| Enrichment Score (ES) | 0.60581577 |
| Normalized Enrichment Score (NES) | 1.6203507 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.23322211 |
| FWER p-Value | 0.618 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS15A | 1453467_s_at 1457726_at | 7 | 4.826 | 0.1431 | Yes | ||
| 2 | UHRF2 | 1447217_at 1454920_at 1457070_at | 17 | 3.839 | 0.2568 | Yes | ||
| 3 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 58 | 2.672 | 0.3343 | Yes | ||
| 4 | SUV39H1 | 1427382_a_at 1432227_at 1432236_a_at | 204 | 1.726 | 0.3790 | Yes | ||
| 5 | MATR3 | 1434888_a_at 1438368_a_at 1441272_at 1450874_at 1458508_at | 360 | 1.291 | 0.4102 | Yes | ||
| 6 | DDX3X | 1416467_at 1423042_at 1423043_s_at | 394 | 1.231 | 0.4453 | Yes | ||
| 7 | THAP7 | 1426428_at 1426429_at 1452069_a_at | 526 | 0.961 | 0.4679 | Yes | ||
| 8 | PLOD2 | 1416686_at 1416687_at | 619 | 0.840 | 0.4886 | Yes | ||
| 9 | HSPA4 | 1416146_at 1416147_at 1440575_at | 667 | 0.780 | 0.5096 | Yes | ||
| 10 | COX11 | 1429188_at 1457846_at | 861 | 0.620 | 0.5192 | Yes | ||
| 11 | UPF3B | 1434842_s_at | 1168 | 0.463 | 0.5190 | Yes | ||
| 12 | POLDIP3 | 1434176_x_at 1437335_x_at 1437837_x_at 1440022_at 1452709_at 1455788_x_at | 1315 | 0.405 | 0.5243 | Yes | ||
| 13 | ZFP91 | 1426326_at 1429615_at 1429616_at 1459444_at | 1425 | 0.371 | 0.5303 | Yes | ||
| 14 | ZZZ3 | 1434332_at 1438487_s_at 1441748_at | 1448 | 0.365 | 0.5402 | Yes | ||
| 15 | THAP1 | 1429282_at 1455003_at | 1528 | 0.348 | 0.5469 | Yes | ||
| 16 | POU4F1 | 1429667_at 1429668_at | 1649 | 0.323 | 0.5510 | Yes | ||
| 17 | PPP1R12A | 1429487_at 1437734_at 1437735_at 1444762_at 1444834_at 1453163_at | 1682 | 0.316 | 0.5589 | Yes | ||
| 18 | CHUK | 1417091_at 1428210_s_at 1451383_a_at | 1709 | 0.311 | 0.5670 | Yes | ||
| 19 | OFD1 | 1427172_at | 1790 | 0.297 | 0.5721 | Yes | ||
| 20 | ASXL1 | 1434561_at 1435077_at 1458380_at | 1886 | 0.278 | 0.5761 | Yes | ||
| 21 | BCAS3 | 1423528_at 1428454_at 1457817_at 1458557_at | 1900 | 0.277 | 0.5837 | Yes | ||
| 22 | ATP5F1 | 1426742_at 1433562_s_at | 1943 | 0.269 | 0.5898 | Yes | ||
| 23 | PIGN | 1421284_at 1425813_at 1425997_a_at 1432115_a_at | 2002 | 0.261 | 0.5949 | Yes | ||
| 24 | CDC5L | 1428092_at 1460429_at | 2040 | 0.255 | 0.6007 | Yes | ||
| 25 | FGFR1OP2 | 1423591_at 1427526_at 1431020_a_at 1441460_at | 2091 | 0.248 | 0.6058 | Yes | ||
| 26 | CD2AP | 1420906_at 1420907_at 1420908_at 1460140_at | 2566 | 0.194 | 0.5899 | No | ||
| 27 | RPL17 | 1423855_x_at 1435791_x_at 1453752_at | 4264 | 0.106 | 0.5153 | No | ||
| 28 | TTC14 | 1426544_a_at 1428846_at 1432130_a_at 1438051_at 1452926_at 1460448_s_at | 5002 | 0.087 | 0.4841 | No | ||
| 29 | RBPSUH | 1418114_at 1438848_at 1446436_at 1448957_at 1454896_at | 5126 | 0.084 | 0.4810 | No | ||
| 30 | RPS6 | 1453466_at | 5418 | 0.078 | 0.4700 | No | ||
| 31 | RAB33B | 1423083_at | 5432 | 0.077 | 0.4717 | No | ||
| 32 | MDH2 | 1416478_a_at 1433984_a_at | 5482 | 0.076 | 0.4717 | No | ||
| 33 | SCAMP5 | 1450247_a_at 1451224_at | 5641 | 0.073 | 0.4666 | No | ||
| 34 | GAS8 | 1418361_at 1440788_at | 5661 | 0.073 | 0.4679 | No | ||
| 35 | ATF7 | 1425653_at 1437645_at 1445509_at | 5689 | 0.072 | 0.4688 | No | ||
| 36 | CNOT3 | 1435965_at 1441465_at | 5746 | 0.071 | 0.4684 | No | ||
| 37 | AGL | 1429083_at 1431032_at 1431033_x_at | 6464 | 0.057 | 0.4373 | No | ||
| 38 | TROAP | 1433790_at 1453370_at | 7070 | 0.047 | 0.4109 | No | ||
| 39 | KAZALD1 | 1436528_at | 7543 | 0.039 | 0.3905 | No | ||
| 40 | NDUFA11 | 1429708_at 1444154_at | 7964 | 0.033 | 0.3722 | No | ||
| 41 | ADNP | 1423069_at | 8145 | 0.031 | 0.3649 | No | ||
| 42 | DBR1 | 1451641_at | 8149 | 0.031 | 0.3657 | No | ||
| 43 | MOBKL1A | 1430564_at 1455184_at | 8171 | 0.030 | 0.3656 | No | ||
| 44 | SMARCD2 | 1426192_at 1448400_a_at 1448401_at | 9082 | 0.018 | 0.3245 | No | ||
| 45 | SSR4 | 1427096_s_at 1435709_at 1448524_s_at | 9680 | 0.010 | 0.2974 | No | ||
| 46 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 9944 | 0.006 | 0.2856 | No | ||
| 47 | LUC7L2 | 1423251_at 1427087_at 1436165_at 1436766_at 1436767_at 1437761_at 1458423_at | 9988 | 0.006 | 0.2838 | No | ||
| 48 | CCNJ | 1455653_at | 10058 | 0.004 | 0.2807 | No | ||
| 49 | LRSAM1 | 1429176_at | 10063 | 0.004 | 0.2807 | No | ||
| 50 | PTPN4 | 1421330_at | 10257 | 0.002 | 0.2719 | No | ||
| 51 | PFKFB2 | 1422090_a_at 1422091_at 1422092_at 1429486_at 1431901_a_at | 10716 | -0.004 | 0.2510 | No | ||
| 52 | PRDX1 | 1416000_a_at 1433866_x_at 1434731_x_at 1436691_x_at | 10947 | -0.007 | 0.2407 | No | ||
| 53 | SLC12A2 | 1417622_at 1417623_at 1443332_at 1448780_at | 11192 | -0.010 | 0.2298 | No | ||
| 54 | RPL10A | 1431177_a_at | 11198 | -0.010 | 0.2299 | No | ||
| 55 | DCTN2 | 1424461_at | 11408 | -0.013 | 0.2207 | No | ||
| 56 | SLC25A11 | 1426586_at | 11457 | -0.014 | 0.2190 | No | ||
| 57 | STXBP4 | 1442267_at 1446648_at 1451063_at 1453989_at 1455706_at | 11839 | -0.019 | 0.2021 | No | ||
| 58 | RPS19 | 1449243_a_at 1460442_at | 11847 | -0.019 | 0.2023 | No | ||
| 59 | BANP | 1452462_a_at | 12023 | -0.021 | 0.1949 | No | ||
| 60 | TAS1R1 | 1420432_at | 12268 | -0.024 | 0.1844 | No | ||
| 61 | NFYC | 1448963_at | 12833 | -0.032 | 0.1596 | No | ||
| 62 | FSIP1 | 1428035_at | 13501 | -0.041 | 0.1302 | No | ||
| 63 | SEC63 | 1419819_s_at 1424924_at 1424925_at 1424926_at 1457988_at | 14201 | -0.052 | 0.0998 | No | ||
| 64 | SUPT5H | 1424255_at | 14362 | -0.055 | 0.0941 | No | ||
| 65 | MRP63 | 1418137_at 1460225_at | 14620 | -0.059 | 0.0840 | No | ||
| 66 | RPL26 | 1436995_a_at 1448109_a_at | 14818 | -0.062 | 0.0769 | No | ||
| 67 | BZW1 | 1423040_at 1423041_a_at 1450845_a_at 1450846_at | 14999 | -0.065 | 0.0706 | No | ||
| 68 | IDH3G | 1416788_a_at 1416789_at | 15141 | -0.067 | 0.0661 | No | ||
| 69 | RPL12 | 1416026_a_at | 15162 | -0.068 | 0.0672 | No | ||
| 70 | CHURC1 | 1425455_a_at | 15358 | -0.071 | 0.0604 | No | ||
| 71 | HNRPK | 1423684_at 1448176_a_at 1454692_x_at 1460547_a_at | 15570 | -0.075 | 0.0529 | No | ||
| 72 | LAMP1 | 1415880_a_at 1460096_at | 15873 | -0.080 | 0.0415 | No | ||
| 73 | DENR | 1420367_at 1420368_at 1446948_at 1449598_at | 16185 | -0.086 | 0.0298 | No | ||
| 74 | RPS7 | 1435593_x_at | 16380 | -0.090 | 0.0236 | No | ||
| 75 | DGUOK | 1425228_a_at | 16446 | -0.092 | 0.0233 | No | ||
| 76 | ZNF346 | 1417088_at 1448581_at | 16911 | -0.102 | 0.0051 | No | ||
| 77 | ICMT | 1426500_at | 17372 | -0.115 | -0.0125 | No | ||
| 78 | NUDT2 | 1418737_at | 17412 | -0.116 | -0.0109 | No | ||
| 79 | EIF5 | 1415723_at 1433631_at 1454663_at 1454664_a_at 1456256_at | 17633 | -0.122 | -0.0173 | No | ||
| 80 | TLK2 | 1416944_a_at 1431827_a_at 1454018_at 1457357_at | 17679 | -0.123 | -0.0157 | No | ||
| 81 | PSMD5 | 1423234_at | 17740 | -0.125 | -0.0148 | No | ||
| 82 | PRDX4 | 1416166_a_at 1416167_at 1431269_at | 17773 | -0.126 | -0.0125 | No | ||
| 83 | SLC25A4 | 1424562_a_at 1424563_at 1434897_a_at 1455069_x_at | 17841 | -0.128 | -0.0118 | No | ||
| 84 | CAMK2D | 1422659_at 1427763_a_at 1439168_at 1444031_at 1444281_at 1446663_at 1456844_at 1459457_at 1459600_at 1460630_at | 18059 | -0.135 | -0.0177 | No | ||
| 85 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18421 | -0.146 | -0.0299 | No | ||
| 86 | VDAC3 | 1416175_a_at | 18551 | -0.151 | -0.0313 | No | ||
| 87 | CCT8 | 1415785_a_at 1436973_at | 18572 | -0.152 | -0.0277 | No | ||
| 88 | SAP18 | 1419443_at 1419444_at 1419445_s_at 1434646_s_at 1449480_at | 18697 | -0.157 | -0.0287 | No | ||
| 89 | TLOC1 | 1432635_a_at 1433704_s_at 1439233_at 1444811_at 1454697_at 1455698_at | 18963 | -0.168 | -0.0359 | No | ||
| 90 | WDR6 | 1415770_at 1455940_x_at | 18970 | -0.168 | -0.0311 | No | ||
| 91 | RRAGA | 1428905_at | 19188 | -0.179 | -0.0358 | No | ||
| 92 | ARF3 | 1421789_s_at 1423973_a_at 1434787_at 1437331_a_at 1456007_at | 19307 | -0.185 | -0.0357 | No | ||
| 93 | MAP3K7 | 1419988_at 1425795_a_at 1426627_at 1449693_at 1455441_at 1458787_at | 19428 | -0.192 | -0.0355 | No | ||
| 94 | ATF2 | 1426582_at 1426583_at 1427559_a_at 1437777_at 1452116_s_at 1459237_at | 20021 | -0.238 | -0.0555 | No | ||
| 95 | FUS | 1451285_at 1451286_s_at 1455831_at 1458801_at | 20535 | -0.301 | -0.0700 | No | ||
| 96 | ZF | 1425698_a_at 1438302_at 1452856_at 1452857_at | 20985 | -0.399 | -0.0788 | No | ||
| 97 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 21187 | -0.479 | -0.0737 | No | ||
| 98 | ANKFY1 | 1417685_at 1443654_at | 21309 | -0.559 | -0.0626 | No | ||
| 99 | ASH2L | 1420615_at 1420616_at | 21313 | -0.562 | -0.0461 | No | ||
| 100 | ANAPC4 | 1423930_at 1423931_s_at | 21652 | -0.970 | -0.0327 | No | ||
| 101 | MBTPS1 | 1431385_a_at 1431386_s_at 1448240_at | 21800 | -1.535 | 0.0061 | No |

