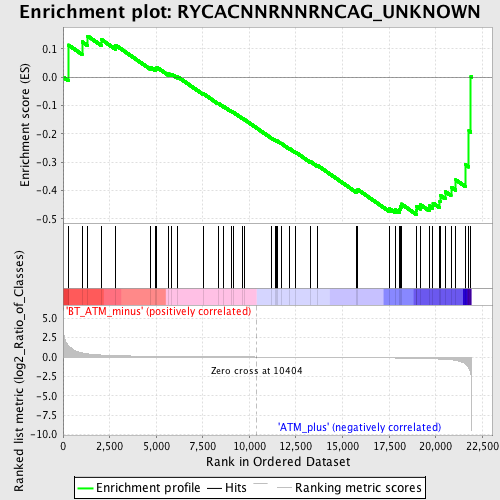
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | RYCACNNRNNRNCAG_UNKNOWN |
| Enrichment Score (ES) | -0.48487246 |
| Normalized Enrichment Score (NES) | -1.3154374 |
| Nominal p-value | 0.10966057 |
| FDR q-value | 0.7099529 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 284 | 1.486 | 0.1134 | No | ||
| 2 | CHRDL1 | 1421295_at 1424945_at 1434201_at 1456722_at | 1022 | 0.525 | 0.1244 | No | ||
| 3 | FBXO11 | 1434108_at | 1325 | 0.401 | 0.1448 | No | ||
| 4 | LDB2 | 1421101_a_at 1439557_s_at 1445126_at 1456786_at | 2059 | 0.252 | 0.1327 | No | ||
| 5 | FGF13 | 1418497_at 1418498_at | 2812 | 0.174 | 0.1132 | No | ||
| 6 | ARX | 1420926_at 1441175_at 1450042_at | 4700 | 0.094 | 0.0350 | No | ||
| 7 | ACCN2 | 1455328_at | 4974 | 0.088 | 0.0300 | No | ||
| 8 | ODF1 | 1422585_at 1440821_x_at 1457659_x_at | 5035 | 0.086 | 0.0346 | No | ||
| 9 | NEUROD2 | 1444362_at | 5636 | 0.073 | 0.0134 | No | ||
| 10 | ITSN2 | 1423184_at 1431772_a_at 1435023_at 1445375_at 1446595_at 1446735_at 1446911_at | 5826 | 0.069 | 0.0106 | No | ||
| 11 | PHLDB1 | 1424467_at 1424468_s_at | 6135 | 0.064 | 0.0020 | No | ||
| 12 | FYB | 1452117_a_at | 7511 | 0.040 | -0.0574 | No | ||
| 13 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 8329 | 0.028 | -0.0924 | No | ||
| 14 | H2AFY | 1424572_a_at | 8625 | 0.024 | -0.1038 | No | ||
| 15 | SEMA3A | 1420416_at 1420417_at 1449865_at | 9035 | 0.018 | -0.1210 | No | ||
| 16 | NPFF | 1420606_at | 9133 | 0.017 | -0.1239 | No | ||
| 17 | CALB1 | 1417504_at 1448738_at 1458836_at | 9624 | 0.010 | -0.1454 | No | ||
| 18 | STARD13 | 1452604_at | 9757 | 0.009 | -0.1507 | No | ||
| 19 | SOX12 | 1439754_at 1455562_at 1456882_at | 11208 | -0.011 | -0.2161 | No | ||
| 20 | MB | 1422420_at 1451203_at | 11378 | -0.013 | -0.2227 | No | ||
| 21 | IL17RE | 1426566_s_at 1439357_at 1452109_at | 11471 | -0.014 | -0.2257 | No | ||
| 22 | WNT10B | 1426091_a_at | 11487 | -0.014 | -0.2252 | No | ||
| 23 | LEMD2 | 1424326_at | 11719 | -0.017 | -0.2343 | No | ||
| 24 | SP8 | 1440519_at | 12177 | -0.023 | -0.2532 | No | ||
| 25 | UPK2 | 1421781_at | 12472 | -0.027 | -0.2644 | No | ||
| 26 | ARTN | 1432032_a_at | 13295 | -0.038 | -0.2987 | No | ||
| 27 | BLOC1S1 | 1422614_s_at | 13660 | -0.044 | -0.3116 | No | ||
| 28 | EMX2 | 1456258_at | 15754 | -0.078 | -0.4006 | No | ||
| 29 | NR5A1 | 1418315_at 1421730_at | 15830 | -0.079 | -0.3973 | No | ||
| 30 | ELMO3 | 1434489_at | 17525 | -0.119 | -0.4646 | No | ||
| 31 | SLC25A4 | 1424562_a_at 1424563_at 1434897_a_at 1455069_x_at | 17841 | -0.128 | -0.4681 | No | ||
| 32 | PBXIP1 | 1451132_at | 18058 | -0.135 | -0.4665 | No | ||
| 33 | CPNE1 | 1433439_at | 18111 | -0.136 | -0.4573 | No | ||
| 34 | PLXNA2 | 1429772_at 1446420_at 1451753_at 1453286_at 1455037_at 1458102_at 1459033_at | 18161 | -0.138 | -0.4478 | No | ||
| 35 | RHOC | 1435394_s_at 1448605_at | 18974 | -0.168 | -0.4705 | Yes | ||
| 36 | CD79B | 1417640_at | 18990 | -0.169 | -0.4568 | Yes | ||
| 37 | RRAGA | 1428905_at | 19188 | -0.179 | -0.4506 | Yes | ||
| 38 | TLE3 | 1419654_at 1419655_at 1449554_at 1458512_at | 19661 | -0.208 | -0.4545 | Yes | ||
| 39 | MDK | 1416006_at | 19862 | -0.223 | -0.4447 | Yes | ||
| 40 | DLGAP4 | 1426465_at 1455548_at | 20232 | -0.262 | -0.4393 | Yes | ||
| 41 | EGR3 | 1421486_at 1436329_at | 20255 | -0.265 | -0.4177 | Yes | ||
| 42 | PPP3CB | 1427468_at 1428473_at 1428474_at 1433835_at 1446149_at 1459378_at | 20516 | -0.299 | -0.4042 | Yes | ||
| 43 | UBE2L3 | 1417907_at 1417908_s_at 1448879_at 1448880_at | 20832 | -0.357 | -0.3882 | Yes | ||
| 44 | PITPNM1 | 1425760_a_at 1437724_x_at 1438854_x_at 1438974_x_at | 21090 | -0.437 | -0.3628 | Yes | ||
| 45 | TPM3 | 1427260_a_at 1427567_a_at 1436958_x_at 1449996_a_at 1449997_at | 21624 | -0.911 | -0.3097 | Yes | ||
| 46 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 21793 | -1.500 | -0.1897 | Yes | ||
| 47 | RHOF | 1434794_at 1441510_at 1455902_x_at | 21901 | -2.305 | 0.0015 | Yes |

