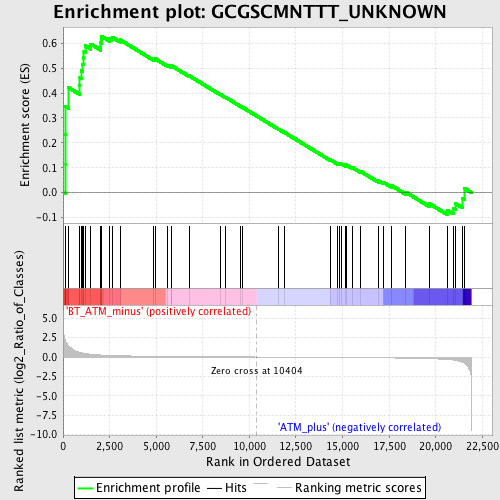
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | GCGSCMNTTT_UNKNOWN |
| Enrichment Score (ES) | 0.6283583 |
| Normalized Enrichment Score (NES) | 1.5379555 |
| Nominal p-value | 0.0224 |
| FDR q-value | 0.2428558 |
| FWER p-Value | 0.945 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADK | 1416319_at 1421767_at 1438292_x_at 1442615_at 1445402_at 1446068_at 1446675_at 1456960_at 1459645_at | 116 | 2.125 | 0.1148 | Yes | ||
| 2 | TARDBP | 1423723_s_at 1428467_at 1434419_s_at 1436318_at 1455655_a_at | 119 | 2.117 | 0.2343 | Yes | ||
| 3 | MYST2 | 1433433_at 1440746_at 1447631_at | 134 | 1.981 | 0.3457 | Yes | ||
| 4 | GABPB2 | 1440289_at 1440868_at 1443942_at 1453682_at 1455110_at | 291 | 1.477 | 0.4220 | Yes | ||
| 5 | SUPT16H | 1419741_at 1449578_at 1456449_at | 875 | 0.606 | 0.4296 | Yes | ||
| 6 | BCLAF1 | 1428844_a_at 1428845_at 1436023_at 1438089_a_at | 902 | 0.590 | 0.4618 | Yes | ||
| 7 | PAICS | 1423564_a_at 1423565_at 1436298_x_at | 981 | 0.549 | 0.4893 | Yes | ||
| 8 | PPAT | 1428543_at 1441770_at 1452831_s_at | 1048 | 0.513 | 0.5152 | Yes | ||
| 9 | STAG2 | 1421849_at 1450396_at | 1081 | 0.498 | 0.5419 | Yes | ||
| 10 | NPAT | 1456485_at 1459185_at | 1119 | 0.485 | 0.5676 | Yes | ||
| 11 | SAFB | 1432911_at 1454415_at 1454928_at 1454929_s_at | 1203 | 0.452 | 0.5893 | Yes | ||
| 12 | GPC3 | 1443240_at 1446525_at 1450990_at | 1488 | 0.355 | 0.5964 | Yes | ||
| 13 | CSE1L | 1448809_at | 2021 | 0.258 | 0.5867 | Yes | ||
| 14 | RAB1A | 1416082_at 1448210_at 1457537_at | 2026 | 0.258 | 0.6011 | Yes | ||
| 15 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 2047 | 0.253 | 0.6145 | Yes | ||
| 16 | RBMX | 1416177_at 1416354_at 1416355_at 1426863_at 1437847_x_at | 2057 | 0.252 | 0.6284 | Yes | ||
| 17 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 2508 | 0.201 | 0.6192 | No | ||
| 18 | FBXL7 | 1439890_at 1440441_at 1446967_at 1456220_at 1457966_at 1458485_at | 2650 | 0.187 | 0.6233 | No | ||
| 19 | ATM | 1421205_at 1428830_at | 3060 | 0.158 | 0.6135 | No | ||
| 20 | APRIN | 1435242_at 1436161_at 1441553_at | 4865 | 0.090 | 0.5362 | No | ||
| 21 | COL27A1 | 1429549_at 1453191_at | 4950 | 0.088 | 0.5373 | No | ||
| 22 | EN1 | 1418618_at | 5623 | 0.073 | 0.5108 | No | ||
| 23 | PITX1 | 1419514_at 1449488_at | 5818 | 0.069 | 0.5058 | No | ||
| 24 | HOXC4 | 1422870_at | 5840 | 0.069 | 0.5088 | No | ||
| 25 | AP4M1 | 1418846_at | 6784 | 0.052 | 0.4686 | No | ||
| 26 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 8436 | 0.026 | 0.3947 | No | ||
| 27 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 8726 | 0.022 | 0.3827 | No | ||
| 28 | NEUROD1 | 1426412_at 1426413_at | 9542 | 0.012 | 0.3461 | No | ||
| 29 | ATOH1 | 1449822_at | 9611 | 0.011 | 0.3436 | No | ||
| 30 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 11574 | -0.015 | 0.2549 | No | ||
| 31 | PSPC1 | 1423192_at 1423193_at 1444748_at 1444957_at 1457723_at | 11902 | -0.019 | 0.2410 | No | ||
| 32 | DLEU2 | 1427410_at 1427411_s_at 1456145_at | 14368 | -0.055 | 0.1315 | No | ||
| 33 | YARS | 1460638_at | 14742 | -0.061 | 0.1179 | No | ||
| 34 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 14844 | -0.062 | 0.1168 | No | ||
| 35 | ARID4A | 1436191_at 1438464_at 1456420_at | 14958 | -0.064 | 0.1153 | No | ||
| 36 | PRDM10 | 1441764_at 1444812_at | 15172 | -0.068 | 0.1094 | No | ||
| 37 | HOXA2 | 1419602_at | 15218 | -0.068 | 0.1112 | No | ||
| 38 | PPP2R2A | 1429715_at 1437730_at 1453260_a_at | 15520 | -0.074 | 0.1016 | No | ||
| 39 | PHF12 | 1434922_at 1453579_at | 15961 | -0.082 | 0.0861 | No | ||
| 40 | NKIRAS2 | 1424416_at | 16930 | -0.103 | 0.0477 | No | ||
| 41 | USP11 | 1426539_at | 17200 | -0.110 | 0.0417 | No | ||
| 42 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 17638 | -0.122 | 0.0286 | No | ||
| 43 | DNAJC7 | 1448362_at 1459704_at | 18399 | -0.145 | 0.0021 | No | ||
| 44 | PPP1R15B | 1436366_at | 19673 | -0.209 | -0.0443 | No | ||
| 45 | CD4 | 1419696_at 1427779_a_at | 20621 | -0.316 | -0.0697 | No | ||
| 46 | HIP2 | 1417186_at 1417187_at 1417188_s_at 1441012_at 1442671_at 1443148_at 1444212_at | 20968 | -0.394 | -0.0632 | No | ||
| 47 | EPC1 | 1418850_at 1429914_at 1440198_at 1440742_at 1442279_at 1443675_at 1458315_at | 21059 | -0.427 | -0.0432 | No | ||
| 48 | SCARA3 | 1427020_at | 21447 | -0.676 | -0.0227 | No | ||
| 49 | SUV39H2 | 1422979_at 1433996_at 1436561_at | 21549 | -0.794 | 0.0176 | No |

