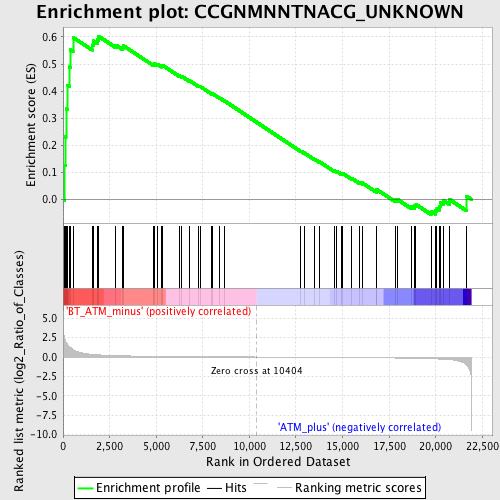
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | CCGNMNNTNACG_UNKNOWN |
| Enrichment Score (ES) | 0.6031778 |
| Normalized Enrichment Score (NES) | 1.5157737 |
| Nominal p-value | 0.022151899 |
| FDR q-value | 0.22505817 |
| FWER p-Value | 0.978 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CYCS | 1422484_at 1445484_at | 92 | 2.381 | 0.1257 | Yes | ||
| 2 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 136 | 1.966 | 0.2310 | Yes | ||
| 3 | DIABLO | 1417683_at 1425768_at | 155 | 1.910 | 0.3344 | Yes | ||
| 4 | BTAF1 | 1435248_a_at 1435249_at 1435953_at | 224 | 1.654 | 0.4215 | Yes | ||
| 5 | CNOT4 | 1420999_at 1421000_at 1436645_a_at 1437586_at 1443169_at 1450083_at 1459449_at | 345 | 1.342 | 0.4892 | Yes | ||
| 6 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 406 | 1.206 | 0.5523 | Yes | ||
| 7 | ZBTB11 | 1454826_at | 533 | 0.952 | 0.5985 | Yes | ||
| 8 | DDX50 | 1417875_at 1439082_at | 1556 | 0.343 | 0.5704 | Yes | ||
| 9 | SYT6 | 1420188_at 1426106_a_at 1449766_at 1449767_x_at | 1622 | 0.328 | 0.5853 | Yes | ||
| 10 | HSPD1 | 1426351_at | 1860 | 0.283 | 0.5899 | Yes | ||
| 11 | BMP2K | 1421103_at 1437419_at 1458370_at | 1901 | 0.276 | 0.6032 | Yes | ||
| 12 | RBM18 | 1420607_at 1420608_at 1458182_at | 2831 | 0.173 | 0.5702 | No | ||
| 13 | SFRS10 | 1419543_a_at 1427432_a_at 1441466_at 1446496_at | 3189 | 0.150 | 0.5621 | No | ||
| 14 | TAF11 | 1446161_at 1451995_at | 3255 | 0.147 | 0.5671 | No | ||
| 15 | HIP1 | 1424755_at 1424756_at 1432017_at 1432230_at 1434557_at 1444873_at | 4866 | 0.090 | 0.4984 | No | ||
| 16 | RFC4 | 1424321_at 1438161_s_at | 4906 | 0.089 | 0.5015 | No | ||
| 17 | CANX | 1415692_s_at 1422845_at 1428935_at | 5074 | 0.085 | 0.4985 | No | ||
| 18 | GRWD1 | 1436887_x_at 1452171_at 1455841_s_at | 5285 | 0.081 | 0.4934 | No | ||
| 19 | HERC1 | 1425378_at 1434060_at 1440437_at 1440903_at | 5359 | 0.079 | 0.4943 | No | ||
| 20 | HSPE1 | 1450668_s_at | 6259 | 0.061 | 0.4566 | No | ||
| 21 | PPT2 | 1418302_at | 6367 | 0.059 | 0.4549 | No | ||
| 22 | GNB2 | 1450623_at | 6760 | 0.052 | 0.4398 | No | ||
| 23 | GPM6B | 1423091_a_at 1425942_a_at 1453514_at | 7278 | 0.043 | 0.4186 | No | ||
| 24 | ELK4 | 1422233_at 1427162_a_at 1447571_at 1450549_s_at | 7351 | 0.042 | 0.4176 | No | ||
| 25 | CCBL1 | 1428151_x_at 1438348_x_at 1446302_at 1452678_a_at | 7980 | 0.033 | 0.3907 | No | ||
| 26 | ZCCHC9 | 1424557_at | 8006 | 0.033 | 0.3914 | No | ||
| 27 | ADCK4 | 1448490_at | 8422 | 0.027 | 0.3738 | No | ||
| 28 | PRKACB | 1420610_at 1420611_at 1457197_at | 8690 | 0.023 | 0.3629 | No | ||
| 29 | CHRD | 1417304_at | 12757 | -0.031 | 0.1787 | No | ||
| 30 | MRPL40 | 1448849_at | 12939 | -0.033 | 0.1723 | No | ||
| 31 | THPO | 1449569_at | 13507 | -0.041 | 0.1486 | No | ||
| 32 | FOSB | 1422134_at | 13754 | -0.045 | 0.1398 | No | ||
| 33 | AP1M1 | 1416307_at 1459832_s_at | 14548 | -0.058 | 0.1068 | No | ||
| 34 | RPS27 | 1415716_a_at 1460699_at | 14701 | -0.060 | 0.1031 | No | ||
| 35 | SDF2 | 1416857_at | 14941 | -0.064 | 0.0957 | No | ||
| 36 | BZW1 | 1423040_at 1423041_a_at 1450845_a_at 1450846_at | 14999 | -0.065 | 0.0966 | No | ||
| 37 | MRRF | 1426364_at | 15475 | -0.073 | 0.0789 | No | ||
| 38 | FBXO8 | 1418510_s_at | 15924 | -0.081 | 0.0629 | No | ||
| 39 | HIRA | 1436241_s_at 1450049_a_at 1450050_at 1459103_at | 16070 | -0.084 | 0.0608 | No | ||
| 40 | SPINK2 | 1454710_at | 16815 | -0.100 | 0.0323 | No | ||
| 41 | TXN2 | 1421557_x_at 1452782_a_at 1455640_a_at | 16821 | -0.100 | 0.0375 | No | ||
| 42 | HMG20B | 1417637_a_at | 17824 | -0.128 | -0.0013 | No | ||
| 43 | ITPKC | 1440024_at 1454755_at | 17970 | -0.132 | -0.0008 | No | ||
| 44 | UBE2H | 1418631_at 1418632_at 1428791_at 1438971_x_at 1447374_at 1459930_at | 18707 | -0.157 | -0.0258 | No | ||
| 45 | RNF44 | 1423532_at | 18863 | -0.164 | -0.0240 | No | ||
| 46 | MARK3 | 1418316_a_at | 18949 | -0.167 | -0.0187 | No | ||
| 47 | MFHAS1 | 1429005_at 1436897_at 1439929_at 1459412_at | 19759 | -0.214 | -0.0440 | No | ||
| 48 | ARHGEF7 | 1424482_at 1449066_a_at | 19985 | -0.235 | -0.0415 | No | ||
| 49 | SPRY2 | 1421656_at 1436584_at | 20076 | -0.244 | -0.0323 | No | ||
| 50 | SUPT6H | 1417628_at 1430458_at | 20231 | -0.262 | -0.0250 | No | ||
| 51 | HTATIP | 1433980_at 1433981_s_at | 20270 | -0.266 | -0.0123 | No | ||
| 52 | WDR1 | 1423054_at 1437591_a_at 1450851_at | 20419 | -0.286 | -0.0034 | No | ||
| 53 | KCNN2 | 1445676_at 1448927_at | 20739 | -0.334 | 0.0002 | No | ||
| 54 | CAD | 1440857_at 1452829_at 1452830_s_at | 21661 | -0.996 | 0.0125 | No |

