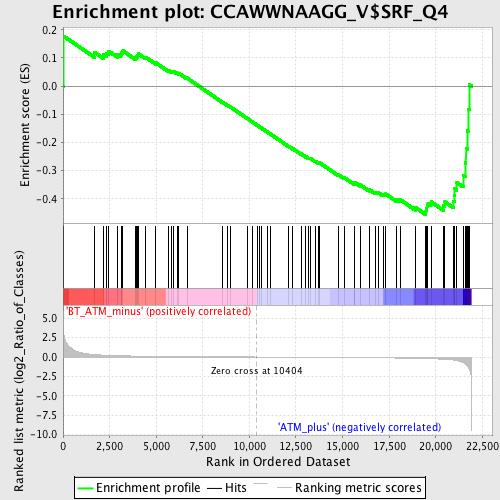
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CCAWWNAAGG_V$SRF_Q4 |
| Enrichment Score (ES) | -0.45561817 |
| Normalized Enrichment Score (NES) | -1.3194886 |
| Nominal p-value | 0.09970675 |
| FDR q-value | 0.7469722 |
| FWER p-Value | 0.999 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ANP32E | 1420592_a_at 1447680_at | 32 | 3.091 | 0.1775 | No | ||
| 2 | PPP1R12A | 1429487_at 1437734_at 1437735_at 1444762_at 1444834_at 1453163_at | 1682 | 0.316 | 0.1203 | No | ||
| 3 | TRDN | 1426134_at 1426142_a_at 1426143_at 1426144_x_at 1451801_at 1451940_x_at | 2179 | 0.238 | 0.1114 | No | ||
| 4 | TCF4 | 1416723_at 1416724_x_at 1416725_at 1424089_a_at 1434148_at 1434149_at 1439336_at 1440106_at 1446386_at 1458201_at | 2341 | 0.219 | 0.1168 | No | ||
| 5 | SCOC | 1416267_at 1430999_a_at | 2442 | 0.207 | 0.1242 | No | ||
| 6 | CORO1C | 1417752_at 1419911_at 1437721_at 1442972_at 1449660_s_at 1459169_at | 2916 | 0.167 | 0.1122 | No | ||
| 7 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 3109 | 0.155 | 0.1124 | No | ||
| 8 | SFRS12 | 1427134_at 1427135_at 1427136_s_at 1458056_at | 3154 | 0.152 | 0.1192 | No | ||
| 9 | RARB | 1454906_at | 3213 | 0.149 | 0.1252 | No | ||
| 10 | ATRX | 1420945_at 1420946_at 1420947_at 1438750_at 1441014_at 1441771_at 1450051_at 1453734_at 1456832_at | 3894 | 0.120 | 0.1010 | No | ||
| 11 | CFL2 | 1418066_at 1418067_at 1431432_at | 3926 | 0.118 | 0.1064 | No | ||
| 12 | SDPR | 1416778_at 1416779_at 1443832_s_at | 3998 | 0.116 | 0.1098 | No | ||
| 13 | UBE2C | 1452954_at | 4036 | 0.115 | 0.1148 | No | ||
| 14 | TPM2 | 1419738_a_at 1419739_at 1425028_a_at 1449577_x_at | 4434 | 0.101 | 0.1025 | No | ||
| 15 | SCN3B | 1426328_a_at 1435767_at | 4975 | 0.088 | 0.0829 | No | ||
| 16 | PICALM | 1441328_at 1443218_at 1446968_at 1451316_a_at 1455773_at | 5637 | 0.073 | 0.0569 | No | ||
| 17 | SRF | 1418255_s_at 1418256_at | 5837 | 0.069 | 0.0518 | No | ||
| 18 | PODN | 1435180_at | 5939 | 0.067 | 0.0510 | No | ||
| 19 | CSRP1 | 1425810_a_at 1425811_a_at | 6115 | 0.064 | 0.0467 | No | ||
| 20 | CKM | 1417614_at | 6219 | 0.062 | 0.0456 | No | ||
| 21 | SP7 | 1418425_at | 6654 | 0.054 | 0.0289 | No | ||
| 22 | MYLK | 1425504_at 1425505_at 1425506_at | 8585 | 0.024 | -0.0580 | No | ||
| 23 | KCTD15 | 1435339_at | 8838 | 0.021 | -0.0683 | No | ||
| 24 | MRVI1 | 1422245_a_at 1459665_s_at | 9005 | 0.019 | -0.0748 | No | ||
| 25 | VIT | 1426231_at | 9915 | 0.006 | -0.1160 | No | ||
| 26 | TAGLN | 1423505_at | 10190 | 0.003 | -0.1283 | No | ||
| 27 | MYH11 | 1418122_at 1448962_at | 10443 | -0.000 | -0.1398 | No | ||
| 28 | PLN | 1423359_at 1450952_at 1460332_at | 10570 | -0.002 | -0.1455 | No | ||
| 29 | MRGPRF | 1425894_at | 10659 | -0.003 | -0.1493 | No | ||
| 30 | TNNC1 | 1418370_at | 10957 | -0.007 | -0.1625 | No | ||
| 31 | KCNK3 | 1425341_at 1425342_a_at 1426058_a_at | 11149 | -0.010 | -0.1707 | No | ||
| 32 | PLCB3 | 1448661_at | 12112 | -0.022 | -0.2134 | No | ||
| 33 | DAPK3 | 1423446_at 1456496_at | 12342 | -0.025 | -0.2224 | No | ||
| 34 | FGFRL1 | 1447878_s_at 1451912_a_at | 12779 | -0.031 | -0.2405 | No | ||
| 35 | EPHB1 | 1455188_at | 13030 | -0.034 | -0.2500 | No | ||
| 36 | NKX2-2 | 1421112_at | 13191 | -0.037 | -0.2552 | No | ||
| 37 | NKX6-1 | 1425828_at | 13267 | -0.038 | -0.2564 | No | ||
| 38 | NPAS2 | 1421036_at 1421037_at | 13539 | -0.042 | -0.2664 | No | ||
| 39 | PFN1 | 1449018_at | 13688 | -0.044 | -0.2706 | No | ||
| 40 | FOSB | 1422134_at | 13754 | -0.045 | -0.2710 | No | ||
| 41 | HOXD10 | 1418606_at | 14797 | -0.062 | -0.3150 | No | ||
| 42 | MYL7 | 1449071_at | 15091 | -0.066 | -0.3246 | No | ||
| 43 | MAPKAPK2 | 1426648_at | 15662 | -0.076 | -0.3463 | No | ||
| 44 | HOXD13 | 1422239_at 1440626_at | 15667 | -0.076 | -0.3420 | No | ||
| 45 | PHF12 | 1434922_at 1453579_at | 15961 | -0.082 | -0.3507 | No | ||
| 46 | INSM1 | 1421399_at 1455865_at | 16436 | -0.092 | -0.3671 | No | ||
| 47 | NPPA | 1456062_at | 16758 | -0.099 | -0.3760 | No | ||
| 48 | DEFB129 | 1440957_at | 16909 | -0.102 | -0.3769 | No | ||
| 49 | HOXC5 | 1439885_at 1450832_at 1456301_at | 17184 | -0.110 | -0.3831 | No | ||
| 50 | ACTG2 | 1422340_a_at | 17319 | -0.113 | -0.3827 | No | ||
| 51 | PRDM1 | 1420425_at 1444997_at | 17890 | -0.129 | -0.4013 | No | ||
| 52 | DUSP5 | 1437199_at | 18110 | -0.136 | -0.4034 | No | ||
| 53 | RAB30 | 1426452_a_at | 18909 | -0.165 | -0.4303 | No | ||
| 54 | LSP1 | 1417756_a_at | 19463 | -0.195 | -0.4444 | Yes | ||
| 55 | RUNDC1 | 1454829_at | 19489 | -0.196 | -0.4341 | Yes | ||
| 56 | ACTA1 | 1427735_a_at | 19541 | -0.199 | -0.4249 | Yes | ||
| 57 | TITF1 | 1422346_at | 19593 | -0.203 | -0.4155 | Yes | ||
| 58 | ERBB2IP | 1428011_a_at 1439079_a_at 1439080_at 1439638_at 1440573_at | 19756 | -0.214 | -0.4105 | Yes | ||
| 59 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 20415 | -0.285 | -0.4241 | Yes | ||
| 60 | DIXDC1 | 1425256_a_at 1435207_at 1443957_at 1444395_at | 20502 | -0.296 | -0.4109 | Yes | ||
| 61 | FLNA | 1426677_at | 20973 | -0.396 | -0.4095 | Yes | ||
| 62 | DUSP2 | 1450698_at | 21013 | -0.410 | -0.3875 | Yes | ||
| 63 | H3F3A | 1423263_at 1442087_at | 21038 | -0.420 | -0.3643 | Yes | ||
| 64 | RASSF2 | 1428392_at 1444889_at | 21144 | -0.460 | -0.3425 | Yes | ||
| 65 | MBNL1 | 1416904_at 1439686_at 1440315_at 1445106_at 1457924_at | 21482 | -0.716 | -0.3164 | Yes | ||
| 66 | NR4A1 | 1416505_at | 21587 | -0.852 | -0.2719 | Yes | ||
| 67 | SVIL | 1427662_at 1460694_s_at 1460735_at | 21635 | -0.928 | -0.2203 | Yes | ||
| 68 | EGR1 | 1417065_at | 21702 | -1.120 | -0.1585 | Yes | ||
| 69 | PCDH7 | 1427592_at 1437442_at 1449249_at 1456214_at | 21768 | -1.369 | -0.0822 | Yes | ||
| 70 | EGR2 | 1427682_a_at 1427683_at | 21804 | -1.550 | 0.0059 | Yes |

