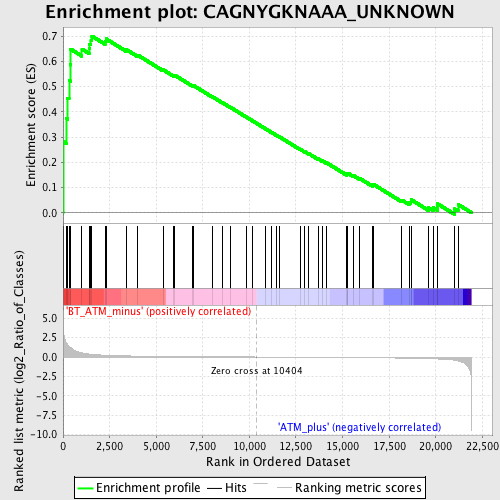
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | CAGNYGKNAAA_UNKNOWN |
| Enrichment Score (ES) | 0.7003778 |
| Normalized Enrichment Score (NES) | 1.7267715 |
| Nominal p-value | 0.0015337423 |
| FDR q-value | 0.16835752 |
| FWER p-Value | 0.155 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 2 | 5.413 | 0.2830 | Yes | ||
| 2 | PPP1CB | 1426205_at 1431328_at 1433540_x_at 1456462_x_at | 164 | 1.886 | 0.3742 | Yes | ||
| 3 | NR3C1 | 1421866_at 1421867_at 1453860_s_at 1457635_s_at 1460303_at | 240 | 1.610 | 0.4550 | Yes | ||
| 4 | APPBP2 | 1430265_at 1434039_at 1434040_at 1434041_at 1444886_at 1447855_x_at 1451251_at | 319 | 1.404 | 0.5249 | Yes | ||
| 5 | SOD2 | 1417193_at 1417194_at 1442994_at 1444531_at 1448610_a_at 1454976_at | 387 | 1.241 | 0.5867 | Yes | ||
| 6 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 406 | 1.206 | 0.6489 | Yes | ||
| 7 | TBL1X | 1434643_at 1434644_at 1441435_at 1446736_at 1455042_at | 1013 | 0.529 | 0.6488 | Yes | ||
| 8 | GFRA1 | 1421973_at 1439015_at 1450440_at | 1396 | 0.380 | 0.6513 | Yes | ||
| 9 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 1432 | 0.370 | 0.6690 | Yes | ||
| 10 | DHX30 | 1416140_a_at 1438832_x_at 1438969_x_at 1453251_at | 1496 | 0.354 | 0.6846 | Yes | ||
| 11 | HNRPD | 1425142_a_at 1446083_at | 1547 | 0.345 | 0.7004 | Yes | ||
| 12 | EIF4A2 | 1450934_at | 2253 | 0.229 | 0.6801 | No | ||
| 13 | RORC | 1425792_a_at 1425793_a_at | 2308 | 0.222 | 0.6893 | No | ||
| 14 | ACVR2B | 1419140_at 1439856_at | 3391 | 0.140 | 0.6472 | No | ||
| 15 | GRIK3 | 1427709_at | 4006 | 0.115 | 0.6252 | No | ||
| 16 | TNFRSF19 | 1415921_a_at 1425212_a_at 1427600_at 1445949_at 1448147_at | 5371 | 0.079 | 0.5669 | No | ||
| 17 | HECTD2 | 1433944_at 1438596_at 1446224_at 1446430_at | 5942 | 0.067 | 0.5444 | No | ||
| 18 | CACNA2D2 | 1426185_at | 5994 | 0.066 | 0.5455 | No | ||
| 19 | PLAC1 | 1417553_at | 6949 | 0.049 | 0.5045 | No | ||
| 20 | BMP1 | 1426238_at 1427457_a_at 1427458_at 1446007_at | 7016 | 0.048 | 0.5040 | No | ||
| 21 | WDTC1 | 1434560_at | 8000 | 0.033 | 0.4608 | No | ||
| 22 | GABRB3 | 1421189_at 1421190_at 1435021_at | 8562 | 0.025 | 0.4365 | No | ||
| 23 | EMILIN3 | 1436965_at | 8968 | 0.019 | 0.4190 | No | ||
| 24 | TRAM1 | 1423732_at 1438225_x_at 1444506_at | 9840 | 0.008 | 0.3796 | No | ||
| 25 | PITX2 | 1424797_a_at 1450482_a_at | 10193 | 0.003 | 0.3636 | No | ||
| 26 | SYTL4 | 1417336_a_at 1425998_at | 10850 | -0.006 | 0.3339 | No | ||
| 27 | BNC2 | 1438861_at 1440593_at 1459132_at | 11214 | -0.011 | 0.3179 | No | ||
| 28 | CRSP8 | 1432462_a_at | 11439 | -0.014 | 0.3084 | No | ||
| 29 | CLSTN2 | 1422158_at 1441165_s_at 1442924_at | 11618 | -0.016 | 0.3011 | No | ||
| 30 | CDH5 | 1422047_at 1433956_at | 12755 | -0.031 | 0.2508 | No | ||
| 31 | GAD2 | 1421978_at | 12976 | -0.034 | 0.2425 | No | ||
| 32 | NKX2-2 | 1421112_at | 13191 | -0.037 | 0.2346 | No | ||
| 33 | HOXA7 | 1449499_at | 13697 | -0.044 | 0.2139 | No | ||
| 34 | CNTN6 | 1422649_at | 13904 | -0.048 | 0.2069 | No | ||
| 35 | RPL13 | 1450150_a_at 1455245_x_at | 14149 | -0.051 | 0.1984 | No | ||
| 36 | RSN | 1425060_s_at 1431098_at 1439800_at 1444750_at 1450351_a_at 1452929_at | 15219 | -0.068 | 0.1532 | No | ||
| 37 | TPI1 | 1415918_a_at 1435659_a_at 1452927_x_at | 15245 | -0.069 | 0.1556 | No | ||
| 38 | MEF2D | 1421388_at 1434487_at 1436642_x_at 1437300_at 1458901_at | 15295 | -0.070 | 0.1570 | No | ||
| 39 | ATP1A2 | 1427465_at 1434893_at 1443823_s_at 1452308_a_at 1455136_at | 15599 | -0.075 | 0.1471 | No | ||
| 40 | RHOBTB1 | 1429206_at 1429656_at | 15915 | -0.081 | 0.1370 | No | ||
| 41 | CER1 | 1450256_at 1450257_at | 16634 | -0.096 | 0.1092 | No | ||
| 42 | TGIF | 1422286_a_at | 16684 | -0.097 | 0.1120 | No | ||
| 43 | PRKACA | 1447720_x_at 1450519_a_at | 18194 | -0.139 | 0.0503 | No | ||
| 44 | SAP30L | 1415718_at 1447379_at 1460071_at | 18583 | -0.152 | 0.0405 | No | ||
| 45 | POFUT1 | 1443881_at 1450137_at 1456323_at | 18681 | -0.156 | 0.0442 | No | ||
| 46 | MYO18A | 1451422_at | 18683 | -0.156 | 0.0524 | No | ||
| 47 | GRK5 | 1446361_at 1446998_at 1449514_at | 19607 | -0.203 | 0.0208 | No | ||
| 48 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 19874 | -0.224 | 0.0204 | No | ||
| 49 | HOXB2 | 1444204_at 1449397_at | 20096 | -0.246 | 0.0232 | No | ||
| 50 | NNAT | 1423506_a_at | 20106 | -0.248 | 0.0358 | No | ||
| 51 | PDE4D | 1443289_at 1458212_at 1459289_at 1459311_at | 21035 | -0.420 | 0.0153 | No | ||
| 52 | AKT2 | 1421324_a_at 1424480_s_at 1455703_at | 21211 | -0.492 | 0.0330 | No |

