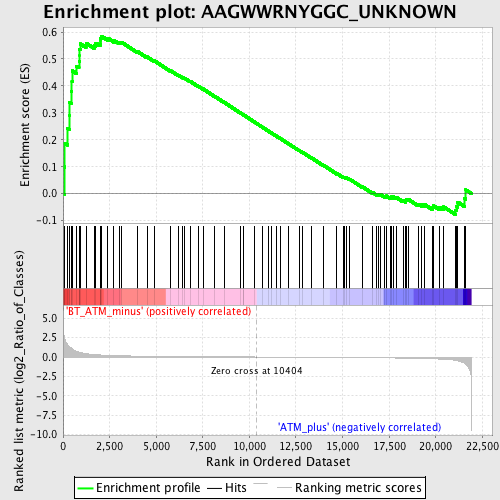
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | AAGWWRNYGGC_UNKNOWN |
| Enrichment Score (ES) | 0.58479357 |
| Normalized Enrichment Score (NES) | 1.5619272 |
| Nominal p-value | 0.011267605 |
| FDR q-value | 0.21523266 |
| FWER p-Value | 0.893 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 58 | 2.672 | 0.0994 | Yes | ||
| 2 | PHF8 | 1439132_at 1459392_at 1460398_at | 94 | 2.348 | 0.1874 | Yes | ||
| 3 | SMYD5 | 1425479_at 1447749_at 1451722_s_at | 247 | 1.592 | 0.2412 | Yes | ||
| 4 | COL4A3BP | 1420383_a_at 1420384_at 1449847_a_at 1452867_at | 337 | 1.361 | 0.2891 | Yes | ||
| 5 | CNOT4 | 1420999_at 1421000_at 1436645_a_at 1437586_at 1443169_at 1450083_at 1459449_at | 345 | 1.342 | 0.3401 | Yes | ||
| 6 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 436 | 1.145 | 0.3797 | Yes | ||
| 7 | ARL1 | 1447089_at 1447090_s_at 1451025_at | 475 | 1.055 | 0.4182 | Yes | ||
| 8 | YY1 | 1422569_at 1422570_at 1435824_at 1457834_at | 496 | 1.017 | 0.4561 | Yes | ||
| 9 | ANKRD28 | 1434630_at 1454801_at | 728 | 0.719 | 0.4730 | Yes | ||
| 10 | PPM1E | 1434990_at 1445741_at | 855 | 0.624 | 0.4911 | Yes | ||
| 11 | SUPT16H | 1419741_at 1449578_at 1456449_at | 875 | 0.606 | 0.5133 | Yes | ||
| 12 | FKRP | 1436812_at 1437536_at 1438414_at | 880 | 0.602 | 0.5362 | Yes | ||
| 13 | RBM3 | 1429169_at | 926 | 0.576 | 0.5561 | Yes | ||
| 14 | ZZEF1 | 1434192_at 1438691_at 1446081_at 1453583_at | 1232 | 0.441 | 0.5589 | Yes | ||
| 15 | EP300 | 1434765_at | 1665 | 0.320 | 0.5514 | Yes | ||
| 16 | HOXA9 | 1421579_at 1455626_at | 1754 | 0.303 | 0.5590 | Yes | ||
| 17 | ATF1 | 1417296_at | 1989 | 0.262 | 0.5582 | Yes | ||
| 18 | UBE3A | 1416680_at 1416681_at 1416682_at 1425206_a_at 1431224_at 1445727_at | 1994 | 0.262 | 0.5681 | Yes | ||
| 19 | PIGN | 1421284_at 1425813_at 1425997_a_at 1432115_a_at | 2002 | 0.261 | 0.5777 | Yes | ||
| 20 | NASP | 1416042_s_at 1416043_at 1440328_at 1442728_at | 2058 | 0.252 | 0.5848 | Yes | ||
| 21 | RALA | 1423137_at 1450870_at 1456049_at | 2405 | 0.212 | 0.5770 | No | ||
| 22 | PPARBP | 1421906_at 1421907_at 1439408_a_at 1446250_at 1447450_at 1448708_at 1450402_at 1453972_x_at | 2731 | 0.180 | 0.5690 | No | ||
| 23 | RNF26 | 1423650_at | 3015 | 0.160 | 0.5622 | No | ||
| 24 | CDC42BPB | 1434652_at 1459167_at | 3152 | 0.153 | 0.5618 | No | ||
| 25 | SNX13 | 1435251_at 1444141_at 1454938_at | 4009 | 0.115 | 0.5270 | No | ||
| 26 | TIA1 | 1416812_at 1416813_at 1416814_at 1423763_x_at 1431708_a_at 1437934_at 1441834_x_at 1453803_at 1454778_x_at | 4516 | 0.099 | 0.5077 | No | ||
| 27 | TSC1 | 1422043_at 1439989_at 1455252_at | 4910 | 0.089 | 0.4931 | No | ||
| 28 | CNOT3 | 1435965_at 1441465_at | 5746 | 0.071 | 0.4576 | No | ||
| 29 | LRRN3 | 1434539_at | 6193 | 0.062 | 0.4396 | No | ||
| 30 | HOXB6 | 1451660_a_at | 6419 | 0.058 | 0.4315 | No | ||
| 31 | POLK | 1419501_at 1449483_at | 6537 | 0.056 | 0.4283 | No | ||
| 32 | ARCN1 | 1423743_at 1436062_at 1444372_at 1451089_a_at | 6835 | 0.051 | 0.4166 | No | ||
| 33 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 7256 | 0.044 | 0.3991 | No | ||
| 34 | RIF1 | 1438516_at 1445487_at | 7514 | 0.040 | 0.3889 | No | ||
| 35 | NRAS | 1422688_a_at 1454060_a_at | 8152 | 0.031 | 0.3609 | No | ||
| 36 | IRAK1 | 1448668_a_at 1460649_at | 8664 | 0.023 | 0.3384 | No | ||
| 37 | NEUROD1 | 1426412_at 1426413_at | 9542 | 0.012 | 0.2987 | No | ||
| 38 | CHCHD7 | 1440158_x_at 1444318_at 1446844_at 1454640_at | 9684 | 0.010 | 0.2926 | No | ||
| 39 | STRN4 | 1426209_at | 9703 | 0.009 | 0.2921 | No | ||
| 40 | RFX3 | 1425413_at 1441253_at 1441702_at | 10301 | 0.001 | 0.2649 | No | ||
| 41 | WDR33 | 1418338_at 1423874_at 1444434_at 1444558_at 1453322_at 1453554_a_at 1455281_at 1456488_at 1456825_at 1458255_at | 10705 | -0.004 | 0.2466 | No | ||
| 42 | SLITRK3 | 1439250_at | 11020 | -0.008 | 0.2325 | No | ||
| 43 | BNC2 | 1438861_at 1440593_at 1459132_at | 11214 | -0.011 | 0.2241 | No | ||
| 44 | CRSP8 | 1432462_a_at | 11439 | -0.014 | 0.2144 | No | ||
| 45 | PSMD8 | 1423296_at | 11687 | -0.017 | 0.2037 | No | ||
| 46 | BCL2L2 | 1423572_at 1430453_a_at 1430454_x_at 1451029_at | 12076 | -0.021 | 0.1868 | No | ||
| 47 | OTX1 | 1437601_at | 12695 | -0.030 | 0.1596 | No | ||
| 48 | NFYC | 1448963_at | 12833 | -0.032 | 0.1546 | No | ||
| 49 | ARF1 | 1420920_a_at 1426390_a_at | 13320 | -0.038 | 0.1338 | No | ||
| 50 | CDKN2C | 1416868_at 1439164_at | 13994 | -0.049 | 0.1049 | No | ||
| 51 | RPS27 | 1415716_a_at 1460699_at | 14701 | -0.060 | 0.0749 | No | ||
| 52 | HOXC10 | 1439798_at | 15057 | -0.066 | 0.0611 | No | ||
| 53 | RAD23A | 1422964_at 1437697_at 1453623_a_at 1455391_at | 15110 | -0.067 | 0.0613 | No | ||
| 54 | HOXA2 | 1419602_at | 15218 | -0.068 | 0.0590 | No | ||
| 55 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 15394 | -0.072 | 0.0537 | No | ||
| 56 | GNAI2 | 1419449_a_at 1435652_a_at | 16084 | -0.084 | 0.0254 | No | ||
| 57 | FMO5 | 1421709_a_at 1440899_at 1450332_s_at | 16611 | -0.095 | 0.0050 | No | ||
| 58 | RHOB | 1449110_at | 16837 | -0.101 | -0.0015 | No | ||
| 59 | NKIRAS2 | 1424416_at | 16930 | -0.103 | -0.0017 | No | ||
| 60 | ANKHD1 | 1419867_a_at 1436597_at 1440754_at 1441597_at 1442641_at 1448008_at 1453023_at 1455759_a_at | 17027 | -0.106 | -0.0021 | No | ||
| 61 | RAB1B | 1424064_at | 17283 | -0.112 | -0.0095 | No | ||
| 62 | TRIM8 | 1418577_at 1427593_at 1438679_at 1460237_at | 17354 | -0.114 | -0.0083 | No | ||
| 63 | APBA1 | 1437940_at 1455369_at 1459605_at | 17583 | -0.121 | -0.0141 | No | ||
| 64 | EIF5 | 1415723_at 1433631_at 1454663_at 1454664_a_at 1456256_at | 17633 | -0.122 | -0.0117 | No | ||
| 65 | CXXC1 | 1452221_a_at 1454106_a_at | 17734 | -0.125 | -0.0115 | No | ||
| 66 | PRDM1 | 1420425_at 1444997_at | 17890 | -0.129 | -0.0137 | No | ||
| 67 | RAB22A | 1424503_at 1424504_at 1451379_at 1454988_s_at | 18259 | -0.140 | -0.0252 | No | ||
| 68 | DNAJC7 | 1448362_at 1459704_at | 18399 | -0.145 | -0.0260 | No | ||
| 69 | SFN | 1448612_at | 18422 | -0.146 | -0.0214 | No | ||
| 70 | MYLIP | 1424988_at | 18537 | -0.151 | -0.0209 | No | ||
| 71 | CLSTN1 | 1421860_at 1421861_at 1445085_at | 19073 | -0.173 | -0.0388 | No | ||
| 72 | MRPL38 | 1430610_at 1444217_at 1447961_s_at | 19237 | -0.181 | -0.0393 | No | ||
| 73 | PLAG1 | 1421745_at 1443943_at | 19384 | -0.189 | -0.0387 | No | ||
| 74 | VAMP2 | 1420833_at 1420834_at | 19854 | -0.222 | -0.0517 | No | ||
| 75 | ADAMTS9 | 1431399_at 1437785_at | 19876 | -0.225 | -0.0441 | No | ||
| 76 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 20223 | -0.261 | -0.0500 | No | ||
| 77 | SFPQ | 1423795_at 1423796_at 1436898_at 1438458_a_at 1438459_x_at 1439058_at | 20442 | -0.288 | -0.0490 | No | ||
| 78 | EPC1 | 1418850_at 1429914_at 1440198_at 1440742_at 1442279_at 1443675_at 1458315_at | 21059 | -0.427 | -0.0608 | No | ||
| 79 | ZBTB4 | 1429722_at 1460488_at | 21116 | -0.450 | -0.0462 | No | ||
| 80 | PPP1R12B | 1431462_at 1434786_at 1441730_at 1457169_at 1457338_at 1460091_at | 21195 | -0.482 | -0.0314 | No | ||
| 81 | SUV39H2 | 1422979_at 1433996_at 1436561_at | 21549 | -0.794 | -0.0172 | No | ||
| 82 | RNF12 | 1417250_at 1418730_at 1418731_at 1439403_x_at 1440850_at 1443167_at | 21625 | -0.911 | 0.0141 | No |

