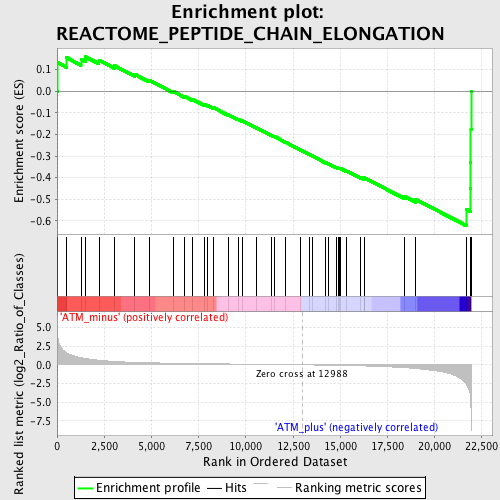
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | REACTOME_PEPTIDE_CHAIN_ELONGATION |
| Enrichment Score (ES) | -0.62373924 |
| Normalized Enrichment Score (NES) | -1.6492193 |
| Nominal p-value | 0.0068259384 |
| FDR q-value | 0.19478102 |
| FWER p-Value | 0.957 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS3 | 1435151_a_at 1447563_at 1455600_at | 12 | 4.863 | 0.1332 | No | ||
| 2 | RPL30 | 1438076_at 1456266_at | 481 | 1.629 | 0.1566 | No | ||
| 3 | RPL4 | 1425183_a_at | 1265 | 0.965 | 0.1473 | No | ||
| 4 | RPS3A | 1422475_a_at | 1510 | 0.847 | 0.1595 | No | ||
| 5 | RPL17 | 1423855_x_at 1435791_x_at 1453752_at | 2218 | 0.607 | 0.1438 | No | ||
| 6 | RPS24 | 1436064_x_at 1436500_at 1453362_x_at 1455195_at 1456628_x_at | 3046 | 0.442 | 0.1182 | No | ||
| 7 | RPS17 | 1437510_x_at 1438501_at 1438986_x_at 1455662_x_at 1457940_at 1459986_a_at | 4106 | 0.328 | 0.0789 | No | ||
| 8 | RPL6 | 1416546_a_at | 4887 | 0.277 | 0.0509 | No | ||
| 9 | RPL8 | 1417762_a_at | 6145 | 0.215 | -0.0007 | No | ||
| 10 | RPL41 | 1422623_x_at 1455578_x_at | 6766 | 0.191 | -0.0237 | No | ||
| 11 | RPS23 | 1460175_at | 7149 | 0.177 | -0.0363 | No | ||
| 12 | RPL11 | 1417615_a_at | 7806 | 0.155 | -0.0620 | No | ||
| 13 | RPL37 | 1453729_a_at | 7946 | 0.150 | -0.0642 | No | ||
| 14 | RPS4X | 1416276_a_at | 8265 | 0.140 | -0.0749 | No | ||
| 15 | RPL27A | 1437729_at | 9071 | 0.115 | -0.1085 | No | ||
| 16 | RPS21 | 1430288_x_at 1433549_x_at 1433721_x_at 1434358_x_at | 9602 | 0.099 | -0.1300 | No | ||
| 17 | RPS2 | 1431766_x_at | 9821 | 0.093 | -0.1374 | No | ||
| 18 | RPL28 | 1416074_a_at | 10571 | 0.072 | -0.1696 | No | ||
| 19 | RPL37A | 1416217_a_at 1460543_x_at | 11375 | 0.049 | -0.2049 | No | ||
| 20 | RPL18A | 1428152_a_at | 11519 | 0.045 | -0.2102 | No | ||
| 21 | RPS5 | 1416054_at | 12081 | 0.027 | -0.2351 | No | ||
| 22 | RPS15 | 1416088_a_at 1416089_at | 12865 | 0.005 | -0.2707 | No | ||
| 23 | RPL19 | 1416219_at | 13348 | -0.012 | -0.2924 | No | ||
| 24 | RPS14 | 1436586_x_at | 13516 | -0.018 | -0.2995 | No | ||
| 25 | RPL35A | 1417317_s_at | 14237 | -0.045 | -0.3312 | No | ||
| 26 | RPS20 | 1456373_x_at 1456436_x_at | 14351 | -0.049 | -0.3350 | No | ||
| 27 | RPLP2 | 1415879_a_at | 14778 | -0.066 | -0.3526 | No | ||
| 28 | RPL18 | 1437005_a_at 1450372_a_at | 14899 | -0.071 | -0.3562 | No | ||
| 29 | RPLP1 | 1416277_a_at | 14964 | -0.074 | -0.3571 | No | ||
| 30 | RPS11 | 1424000_a_at | 15027 | -0.077 | -0.3578 | No | ||
| 31 | RPS28 | 1451101_a_at | 15336 | -0.093 | -0.3693 | No | ||
| 32 | RPL14 | 1422128_at 1426793_a_at 1433688_x_at 1436688_x_at 1438507_x_at 1438626_x_at | 16061 | -0.132 | -0.3987 | No | ||
| 33 | FAU | 1417452_a_at | 16282 | -0.147 | -0.4048 | No | ||
| 34 | RPL39 | 1423032_at 1450840_a_at | 16285 | -0.147 | -0.4008 | No | ||
| 35 | RPL5 | 1423665_a_at 1447460_at | 18408 | -0.380 | -0.4873 | No | ||
| 36 | RPSA | 1416196_at 1448245_at | 18977 | -0.494 | -0.4996 | No | ||
| 37 | EEF2 | 1424736_at 1440784_at 1441414_at | 21695 | -2.721 | -0.5489 | Yes | ||
| 38 | RPS15A | 1453467_s_at 1457726_at | 21880 | -3.935 | -0.4491 | Yes | ||
| 39 | RPS25 | 1430978_at 1451068_s_at | 21895 | -4.317 | -0.3311 | Yes | ||
| 40 | RPS19 | 1449243_a_at 1460442_at | 21918 | -5.655 | -0.1766 | Yes | ||
| 41 | RPS16 | 1416404_s_at 1437196_x_at 1455835_at | 21928 | -6.448 | 0.0003 | Yes |

