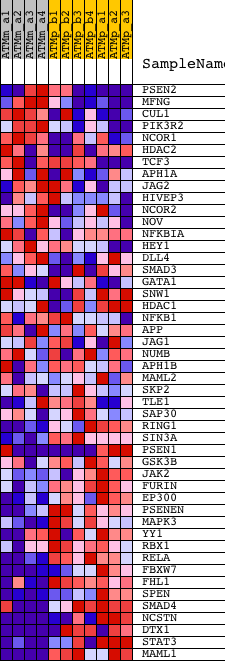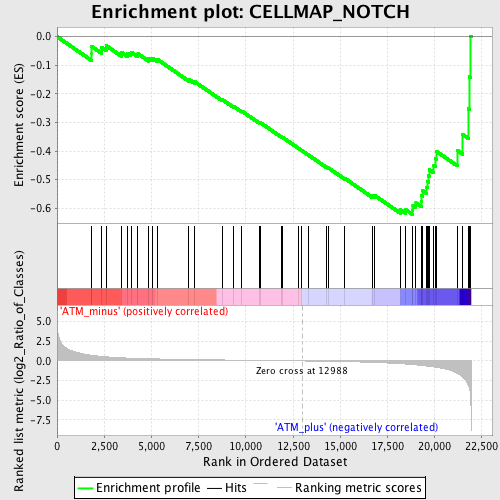
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CELLMAP_NOTCH |
| Enrichment Score (ES) | -0.6216418 |
| Normalized Enrichment Score (NES) | -1.6949823 |
| Nominal p-value | 0.0071174377 |
| FDR q-value | 0.16888118 |
| FWER p-Value | 0.833 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSEN2 | 1425869_a_at 1431542_at | 1813 | 0.728 | -0.0581 | No | ||
| 2 | MFNG | 1416992_at | 1816 | 0.726 | -0.0334 | No | ||
| 3 | CUL1 | 1448174_at 1459516_at | 2342 | 0.582 | -0.0376 | No | ||
| 4 | PIK3R2 | 1418463_at | 2595 | 0.525 | -0.0312 | No | ||
| 5 | NCOR1 | 1423200_at 1423201_at 1423202_a_at 1435914_at 1438276_at 1444760_at 1445230_at 1446574_at 1457007_at 1457938_at 1458103_at | 3418 | 0.396 | -0.0553 | No | ||
| 6 | HDAC2 | 1439704_at 1445684_s_at 1449080_at | 3734 | 0.361 | -0.0574 | No | ||
| 7 | TCF3 | 1436207_at 1450117_at | 3912 | 0.345 | -0.0537 | No | ||
| 8 | APH1A | 1424979_at 1424980_s_at 1451554_a_at | 4281 | 0.315 | -0.0598 | No | ||
| 9 | JAG2 | 1426430_at 1426431_at | 4841 | 0.279 | -0.0759 | No | ||
| 10 | HIVEP3 | 1421150_at 1429134_at 1439660_at 1450132_at 1458802_at | 5038 | 0.267 | -0.0757 | No | ||
| 11 | NCOR2 | 1448893_at 1451841_a_at | 5293 | 0.255 | -0.0787 | No | ||
| 12 | NOV | 1426851_a_at 1426852_x_at | 6976 | 0.183 | -0.1493 | No | ||
| 13 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 7278 | 0.173 | -0.1571 | No | ||
| 14 | HEY1 | 1415999_at | 8748 | 0.125 | -0.2200 | No | ||
| 15 | DLL4 | 1421826_at 1421827_at | 9350 | 0.107 | -0.2438 | No | ||
| 16 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 9779 | 0.094 | -0.2601 | No | ||
| 17 | GATA1 | 1449232_at | 10714 | 0.067 | -0.3005 | No | ||
| 18 | SNW1 | 1429002_at 1429003_at 1459026_at | 10760 | 0.066 | -0.3003 | No | ||
| 19 | HDAC1 | 1448246_at | 11905 | 0.033 | -0.3515 | No | ||
| 20 | NFKB1 | 1427705_a_at 1442949_at | 11951 | 0.032 | -0.3525 | No | ||
| 21 | APP | 1420621_a_at 1427442_a_at 1438373_at 1438374_x_at 1444705_at 1447347_at 1458525_at | 12789 | 0.007 | -0.3905 | No | ||
| 22 | JAG1 | 1421105_at 1421106_at 1434070_at | 12925 | 0.002 | -0.3966 | No | ||
| 23 | NUMB | 1416891_at 1425368_a_at 1444337_at 1457175_at | 13287 | -0.010 | -0.4127 | No | ||
| 24 | APH1B | 1435793_at 1456500_at | 14260 | -0.046 | -0.4556 | No | ||
| 25 | MAML2 | 1446570_at 1447546_s_at | 14370 | -0.050 | -0.4588 | No | ||
| 26 | SKP2 | 1418969_at 1425072_at 1436000_a_at 1437033_a_at 1449293_a_at 1460247_a_at | 15229 | -0.087 | -0.4951 | No | ||
| 27 | TLE1 | 1422751_at 1434033_at | 16683 | -0.179 | -0.5554 | No | ||
| 28 | SAP30 | 1417719_at | 16817 | -0.189 | -0.5550 | No | ||
| 29 | RING1 | 1422647_at | 18170 | -0.348 | -0.6049 | No | ||
| 30 | SIN3A | 1419101_at 1419102_at 1419900_at 1449343_s_at | 18455 | -0.386 | -0.6047 | No | ||
| 31 | PSEN1 | 1420261_at 1421853_at 1425549_at 1450399_at | 18826 | -0.459 | -0.6060 | Yes | ||
| 32 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 18844 | -0.462 | -0.5910 | Yes | ||
| 33 | JAK2 | 1421065_at 1421066_at | 19004 | -0.499 | -0.5813 | Yes | ||
| 34 | FURIN | 1418518_at | 19288 | -0.570 | -0.5748 | Yes | ||
| 35 | EP300 | 1434765_at | 19298 | -0.572 | -0.5557 | Yes | ||
| 36 | PSENEN | 1415679_at | 19351 | -0.584 | -0.5382 | Yes | ||
| 37 | MAPK3 | 1427060_at | 19557 | -0.635 | -0.5260 | Yes | ||
| 38 | YY1 | 1422569_at 1422570_at 1435824_at 1457834_at | 19621 | -0.651 | -0.5067 | Yes | ||
| 39 | RBX1 | 1416577_a_at 1416578_at 1438623_x_at 1440061_at 1447793_x_at 1448387_at | 19670 | -0.667 | -0.4862 | Yes | ||
| 40 | RELA | 1419536_a_at | 19723 | -0.683 | -0.4653 | Yes | ||
| 41 | FBXW7 | 1424986_s_at 1451558_at | 19952 | -0.763 | -0.4497 | Yes | ||
| 42 | FHL1 | 1417872_at 1459003_at | 20044 | -0.797 | -0.4267 | Yes | ||
| 43 | SPEN | 1420397_a_at | 20077 | -0.811 | -0.4006 | Yes | ||
| 44 | SMAD4 | 1422485_at 1422486_a_at 1422487_at 1444205_at | 21192 | -1.576 | -0.3978 | Yes | ||
| 45 | NCSTN | 1418570_at 1444532_at | 21471 | -2.025 | -0.3415 | Yes | ||
| 46 | DTX1 | 1425822_a_at 1458643_at | 21771 | -3.048 | -0.2514 | Yes | ||
| 47 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 21823 | -3.342 | -0.1399 | Yes | ||
| 48 | MAML1 | 1426769_s_at 1441697_at 1445045_at 1452188_at | 21892 | -4.254 | 0.0019 | Yes |