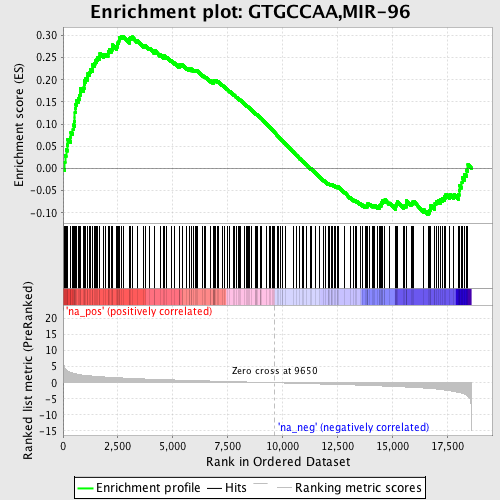
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_LMproB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | GTGCCAA,MIR-96 |
| Enrichment Score (ES) | 0.2983734 |
| Normalized Enrichment Score (NES) | 1.4401659 |
| Nominal p-value | 0.002352941 |
| FDR q-value | 0.7690259 |
| FWER p-Value | 0.999 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | UBE2G1 | 77 | 4.708 | 0.0133 | Yes | ||
| 2 | TRIB3 | 88 | 4.512 | 0.0295 | Yes | ||
| 3 | SLC35A1 | 134 | 4.097 | 0.0423 | Yes | ||
| 4 | PPP3R1 | 190 | 3.765 | 0.0533 | Yes | ||
| 5 | BTBD3 | 212 | 3.691 | 0.0659 | Yes | ||
| 6 | FOXO1A | 347 | 3.220 | 0.0706 | Yes | ||
| 7 | TGFBR1 | 354 | 3.193 | 0.0821 | Yes | ||
| 8 | RAB8B | 449 | 2.974 | 0.0881 | Yes | ||
| 9 | AXUD1 | 465 | 2.951 | 0.0982 | Yes | ||
| 10 | RHPN2 | 526 | 2.859 | 0.1056 | Yes | ||
| 11 | CUGBP2 | 539 | 2.833 | 0.1155 | Yes | ||
| 12 | ARHGAP24 | 540 | 2.832 | 0.1260 | Yes | ||
| 13 | KLHL7 | 552 | 2.806 | 0.1358 | Yes | ||
| 14 | RGS2 | 577 | 2.765 | 0.1448 | Yes | ||
| 15 | LCP1 | 619 | 2.703 | 0.1526 | Yes | ||
| 16 | SORT1 | 689 | 2.618 | 0.1586 | Yes | ||
| 17 | SLC39A1 | 741 | 2.544 | 0.1653 | Yes | ||
| 18 | ABCD1 | 776 | 2.510 | 0.1728 | Yes | ||
| 19 | GGA2 | 797 | 2.493 | 0.1809 | Yes | ||
| 20 | ACE | 947 | 2.338 | 0.1815 | Yes | ||
| 21 | ZFP36L1 | 960 | 2.328 | 0.1895 | Yes | ||
| 22 | KLF3 | 969 | 2.322 | 0.1977 | Yes | ||
| 23 | ZCCHC3 | 1033 | 2.270 | 0.2027 | Yes | ||
| 24 | MAB21L2 | 1101 | 2.223 | 0.2074 | Yes | ||
| 25 | TM9SF4 | 1129 | 2.202 | 0.2141 | Yes | ||
| 26 | VAT1 | 1218 | 2.146 | 0.2173 | Yes | ||
| 27 | CD164 | 1255 | 2.125 | 0.2232 | Yes | ||
| 28 | GPRC5B | 1337 | 2.075 | 0.2265 | Yes | ||
| 29 | VAMP8 | 1342 | 2.072 | 0.2340 | Yes | ||
| 30 | INSIG2 | 1437 | 2.009 | 0.2364 | Yes | ||
| 31 | LAMP2 | 1455 | 1.993 | 0.2428 | Yes | ||
| 32 | SOX6 | 1538 | 1.950 | 0.2456 | Yes | ||
| 33 | ABAT | 1580 | 1.928 | 0.2506 | Yes | ||
| 34 | MTMR12 | 1665 | 1.884 | 0.2530 | Yes | ||
| 35 | MITF | 1677 | 1.875 | 0.2594 | Yes | ||
| 36 | FBXW11 | 1852 | 1.799 | 0.2566 | Yes | ||
| 37 | MORF4L1 | 1948 | 1.748 | 0.2579 | Yes | ||
| 38 | BIRC4 | 2048 | 1.706 | 0.2589 | Yes | ||
| 39 | SLC25A25 | 2058 | 1.701 | 0.2647 | Yes | ||
| 40 | SRCRB4D | 2113 | 1.679 | 0.2680 | Yes | ||
| 41 | DLGAP2 | 2220 | 1.641 | 0.2684 | Yes | ||
| 42 | AP3S1 | 2247 | 1.632 | 0.2730 | Yes | ||
| 43 | SOX5 | 2253 | 1.629 | 0.2788 | Yes | ||
| 44 | PRRX1 | 2444 | 1.565 | 0.2743 | Yes | ||
| 45 | DLAT | 2462 | 1.557 | 0.2792 | Yes | ||
| 46 | TMEPAI | 2498 | 1.547 | 0.2830 | Yes | ||
| 47 | FLOT2 | 2543 | 1.532 | 0.2863 | Yes | ||
| 48 | SLC18A3 | 2549 | 1.530 | 0.2917 | Yes | ||
| 49 | PLAGL1 | 2581 | 1.522 | 0.2957 | Yes | ||
| 50 | PAK1 | 2648 | 1.501 | 0.2977 | Yes | ||
| 51 | SLC10A3 | 2737 | 1.470 | 0.2984 | Yes | ||
| 52 | CNNM3 | 3030 | 1.371 | 0.2876 | No | ||
| 53 | HDAC9 | 3031 | 1.371 | 0.2927 | No | ||
| 54 | ST7 | 3079 | 1.357 | 0.2952 | No | ||
| 55 | ZCCHC11 | 3143 | 1.335 | 0.2967 | No | ||
| 56 | ARPP-19 | 3378 | 1.264 | 0.2887 | No | ||
| 57 | TBX15 | 3666 | 1.177 | 0.2775 | No | ||
| 58 | CTDSP1 | 3757 | 1.153 | 0.2769 | No | ||
| 59 | SLC7A8 | 3937 | 1.107 | 0.2713 | No | ||
| 60 | PLCB4 | 4178 | 1.052 | 0.2621 | No | ||
| 61 | NR4A3 | 4185 | 1.050 | 0.2657 | No | ||
| 62 | SLC22A5 | 4425 | 0.999 | 0.2564 | No | ||
| 63 | ACVR2A | 4585 | 0.962 | 0.2514 | No | ||
| 64 | RAB35 | 4599 | 0.959 | 0.2542 | No | ||
| 65 | TBX1 | 4732 | 0.930 | 0.2505 | No | ||
| 66 | JMJD1C | 4922 | 0.886 | 0.2435 | No | ||
| 67 | CASP2 | 5081 | 0.857 | 0.2381 | No | ||
| 68 | MAP3K3 | 5282 | 0.815 | 0.2303 | No | ||
| 69 | ALK | 5320 | 0.807 | 0.2313 | No | ||
| 70 | CRKL | 5321 | 0.807 | 0.2343 | No | ||
| 71 | NRXN1 | 5421 | 0.784 | 0.2318 | No | ||
| 72 | SNX16 | 5441 | 0.780 | 0.2337 | No | ||
| 73 | PTP4A1 | 5629 | 0.742 | 0.2263 | No | ||
| 74 | NTN4 | 5748 | 0.713 | 0.2225 | No | ||
| 75 | MAP2K1 | 5762 | 0.712 | 0.2244 | No | ||
| 76 | HSPA2 | 5849 | 0.695 | 0.2223 | No | ||
| 77 | GPR124 | 5857 | 0.693 | 0.2245 | No | ||
| 78 | PTGER3 | 5956 | 0.673 | 0.2217 | No | ||
| 79 | NHLH2 | 6015 | 0.663 | 0.2210 | No | ||
| 80 | PPP1R12C | 6063 | 0.652 | 0.2209 | No | ||
| 81 | EXT1 | 6119 | 0.643 | 0.2203 | No | ||
| 82 | LRCH2 | 6353 | 0.599 | 0.2099 | No | ||
| 83 | RNF139 | 6436 | 0.587 | 0.2076 | No | ||
| 84 | STARD7 | 6504 | 0.573 | 0.2061 | No | ||
| 85 | TBL1X | 6714 | 0.535 | 0.1967 | No | ||
| 86 | GRM1 | 6728 | 0.532 | 0.1980 | No | ||
| 87 | PRKAR1A | 6843 | 0.511 | 0.1937 | No | ||
| 88 | STK35 | 6848 | 0.510 | 0.1953 | No | ||
| 89 | AZIN1 | 6849 | 0.510 | 0.1972 | No | ||
| 90 | OVOL1 | 6893 | 0.499 | 0.1968 | No | ||
| 91 | CELSR2 | 6912 | 0.494 | 0.1976 | No | ||
| 92 | PTPN9 | 6914 | 0.494 | 0.1994 | No | ||
| 93 | COBL | 6957 | 0.484 | 0.1989 | No | ||
| 94 | KLK15 | 7042 | 0.468 | 0.1961 | No | ||
| 95 | SLC16A7 | 7069 | 0.463 | 0.1964 | No | ||
| 96 | EPHA3 | 7255 | 0.424 | 0.1879 | No | ||
| 97 | ZHX2 | 7345 | 0.411 | 0.1846 | No | ||
| 98 | TAC1 | 7473 | 0.388 | 0.1792 | No | ||
| 99 | CDC42BPB | 7600 | 0.365 | 0.1737 | No | ||
| 100 | NANOS1 | 7780 | 0.334 | 0.1652 | No | ||
| 101 | ATG16L1 | 7794 | 0.331 | 0.1657 | No | ||
| 102 | BACH2 | 7917 | 0.308 | 0.1602 | No | ||
| 103 | ARID5B | 8012 | 0.289 | 0.1562 | No | ||
| 104 | PPM1F | 8060 | 0.281 | 0.1547 | No | ||
| 105 | CACNB4 | 8103 | 0.275 | 0.1534 | No | ||
| 106 | MYRIP | 8288 | 0.242 | 0.1443 | No | ||
| 107 | TLL1 | 8341 | 0.233 | 0.1423 | No | ||
| 108 | CABP1 | 8409 | 0.224 | 0.1395 | No | ||
| 109 | DHCR24 | 8438 | 0.219 | 0.1388 | No | ||
| 110 | CDK5R2 | 8509 | 0.206 | 0.1358 | No | ||
| 111 | CNN3 | 8602 | 0.189 | 0.1315 | No | ||
| 112 | ST3GAL3 | 8763 | 0.158 | 0.1234 | No | ||
| 113 | SP7 | 8775 | 0.154 | 0.1234 | No | ||
| 114 | PRKCE | 8821 | 0.148 | 0.1215 | No | ||
| 115 | NPTX2 | 8823 | 0.148 | 0.1220 | No | ||
| 116 | BRUNOL6 | 8861 | 0.141 | 0.1205 | No | ||
| 117 | PLSCR3 | 8973 | 0.122 | 0.1149 | No | ||
| 118 | COL13A1 | 8975 | 0.122 | 0.1153 | No | ||
| 119 | MSN | 8997 | 0.117 | 0.1146 | No | ||
| 120 | PDLIM7 | 9042 | 0.110 | 0.1126 | No | ||
| 121 | EBF3 | 9278 | 0.070 | 0.1001 | No | ||
| 122 | GPM6B | 9424 | 0.046 | 0.0924 | No | ||
| 123 | SLC9A2 | 9429 | 0.046 | 0.0923 | No | ||
| 124 | FOXF2 | 9546 | 0.023 | 0.0861 | No | ||
| 125 | SYPL2 | 9595 | 0.011 | 0.0835 | No | ||
| 126 | KPNB1 | 9655 | -0.002 | 0.0803 | No | ||
| 127 | SDC2 | 9769 | -0.024 | 0.0743 | No | ||
| 128 | SH3BP5 | 9823 | -0.036 | 0.0716 | No | ||
| 129 | NLGN2 | 9913 | -0.052 | 0.0669 | No | ||
| 130 | TLOC1 | 9990 | -0.066 | 0.0630 | No | ||
| 131 | FOXQ1 | 9995 | -0.067 | 0.0630 | No | ||
| 132 | SLC38A4 | 10115 | -0.088 | 0.0569 | No | ||
| 133 | LAMC1 | 10477 | -0.153 | 0.0379 | No | ||
| 134 | ITPR2 | 10625 | -0.180 | 0.0305 | No | ||
| 135 | HBEGF | 10795 | -0.209 | 0.0221 | No | ||
| 136 | BRWD1 | 10927 | -0.233 | 0.0159 | No | ||
| 137 | PAPPA | 10978 | -0.242 | 0.0141 | No | ||
| 138 | GPM6A | 11075 | -0.259 | 0.0098 | No | ||
| 139 | ATXN1 | 11290 | -0.304 | -0.0007 | No | ||
| 140 | CSF1 | 11302 | -0.306 | -0.0002 | No | ||
| 141 | CHST10 | 11487 | -0.340 | -0.0089 | No | ||
| 142 | RIMS4 | 11688 | -0.384 | -0.0183 | No | ||
| 143 | PLOD2 | 11872 | -0.418 | -0.0267 | No | ||
| 144 | BAI3 | 11939 | -0.431 | -0.0287 | No | ||
| 145 | ZIC2 | 12079 | -0.456 | -0.0346 | No | ||
| 146 | PPP1R9B | 12113 | -0.462 | -0.0346 | No | ||
| 147 | GPC3 | 12152 | -0.470 | -0.0350 | No | ||
| 148 | KRAS | 12215 | -0.484 | -0.0365 | No | ||
| 149 | SPAST | 12220 | -0.485 | -0.0350 | No | ||
| 150 | TEGT | 12270 | -0.495 | -0.0358 | No | ||
| 151 | BTBD14B | 12353 | -0.510 | -0.0383 | No | ||
| 152 | ZFHX4 | 12427 | -0.524 | -0.0404 | No | ||
| 153 | DDAH1 | 12495 | -0.537 | -0.0420 | No | ||
| 154 | SLC39A10 | 12505 | -0.540 | -0.0405 | No | ||
| 155 | E2F5 | 12555 | -0.549 | -0.0411 | No | ||
| 156 | CAV1 | 12818 | -0.602 | -0.0531 | No | ||
| 157 | MTSS1 | 13094 | -0.659 | -0.0656 | No | ||
| 158 | FARP1 | 13238 | -0.690 | -0.0708 | No | ||
| 159 | SEMA6A | 13317 | -0.707 | -0.0724 | No | ||
| 160 | COL9A1 | 13383 | -0.721 | -0.0733 | No | ||
| 161 | MLLT7 | 13540 | -0.756 | -0.0789 | No | ||
| 162 | ZHX1 | 13651 | -0.782 | -0.0820 | No | ||
| 163 | TPM1 | 13774 | -0.814 | -0.0856 | No | ||
| 164 | EDEM1 | 13806 | -0.822 | -0.0843 | No | ||
| 165 | RPS6KB2 | 13854 | -0.832 | -0.0837 | No | ||
| 166 | SPEN | 13887 | -0.839 | -0.0823 | No | ||
| 167 | CHST1 | 13890 | -0.840 | -0.0793 | No | ||
| 168 | MAP2K3 | 13964 | -0.857 | -0.0801 | No | ||
| 169 | SLC12A5 | 14090 | -0.891 | -0.0836 | No | ||
| 170 | SHC1 | 14142 | -0.904 | -0.0830 | No | ||
| 171 | EIF5 | 14214 | -0.922 | -0.0834 | No | ||
| 172 | LRIG1 | 14350 | -0.961 | -0.0872 | No | ||
| 173 | CACNA2D2 | 14411 | -0.977 | -0.0868 | No | ||
| 174 | CYGB | 14423 | -0.980 | -0.0838 | No | ||
| 175 | ARHGEF12 | 14459 | -0.990 | -0.0820 | No | ||
| 176 | LDB2 | 14508 | -1.004 | -0.0809 | No | ||
| 177 | MECP2 | 14511 | -1.004 | -0.0773 | No | ||
| 178 | ABCA1 | 14551 | -1.018 | -0.0756 | No | ||
| 179 | IRS1 | 14567 | -1.020 | -0.0726 | No | ||
| 180 | PROSC | 14642 | -1.037 | -0.0728 | No | ||
| 181 | MFN1 | 14669 | -1.045 | -0.0703 | No | ||
| 182 | CACNA1C | 14881 | -1.103 | -0.0777 | No | ||
| 183 | CREB3L2 | 15158 | -1.180 | -0.0883 | No | ||
| 184 | TRIM46 | 15163 | -1.182 | -0.0841 | No | ||
| 185 | EIF4EBP2 | 15175 | -1.184 | -0.0803 | No | ||
| 186 | ANKRD27 | 15214 | -1.193 | -0.0780 | No | ||
| 187 | TTYH3 | 15221 | -1.196 | -0.0738 | No | ||
| 188 | AKAP1 | 15514 | -1.290 | -0.0849 | No | ||
| 189 | KCTD16 | 15575 | -1.315 | -0.0833 | No | ||
| 190 | FRAP1 | 15641 | -1.335 | -0.0819 | No | ||
| 191 | RAPGEF4 | 15647 | -1.337 | -0.0772 | No | ||
| 192 | ABCA2 | 15657 | -1.340 | -0.0727 | No | ||
| 193 | RAB23 | 15863 | -1.413 | -0.0786 | No | ||
| 194 | INPP5A | 15904 | -1.433 | -0.0754 | No | ||
| 195 | MTMR3 | 15990 | -1.466 | -0.0746 | No | ||
| 196 | MORF4L2 | 16432 | -1.649 | -0.0924 | No | ||
| 197 | GALNT2 | 16639 | -1.751 | -0.0971 | No | ||
| 198 | SH3KBP1 | 16703 | -1.786 | -0.0939 | No | ||
| 199 | HOOK3 | 16738 | -1.806 | -0.0890 | No | ||
| 200 | SLC1A1 | 16760 | -1.816 | -0.0834 | No | ||
| 201 | HOXA5 | 16923 | -1.908 | -0.0851 | No | ||
| 202 | VIL2 | 16946 | -1.924 | -0.0792 | No | ||
| 203 | LDB1 | 16997 | -1.951 | -0.0746 | No | ||
| 204 | RUFY1 | 17096 | -2.031 | -0.0724 | No | ||
| 205 | SLC1A2 | 17205 | -2.127 | -0.0704 | No | ||
| 206 | RALGPS1 | 17291 | -2.192 | -0.0669 | No | ||
| 207 | OGT | 17382 | -2.270 | -0.0633 | No | ||
| 208 | SLC25A1 | 17446 | -2.330 | -0.0581 | No | ||
| 209 | ADCY6 | 17629 | -2.514 | -0.0586 | No | ||
| 210 | MAGI1 | 17814 | -2.731 | -0.0585 | No | ||
| 211 | LUZP1 | 18029 | -3.008 | -0.0589 | No | ||
| 212 | TNS3 | 18047 | -3.035 | -0.0486 | No | ||
| 213 | RYK | 18073 | -3.070 | -0.0385 | No | ||
| 214 | SLC6A9 | 18177 | -3.263 | -0.0320 | No | ||
| 215 | CELSR1 | 18187 | -3.272 | -0.0203 | No | ||
| 216 | FYN | 18302 | -3.589 | -0.0132 | No | ||
| 217 | CACNB1 | 18389 | -3.913 | -0.0033 | No | ||
| 218 | BCR | 18450 | -4.194 | 0.0090 | No |
