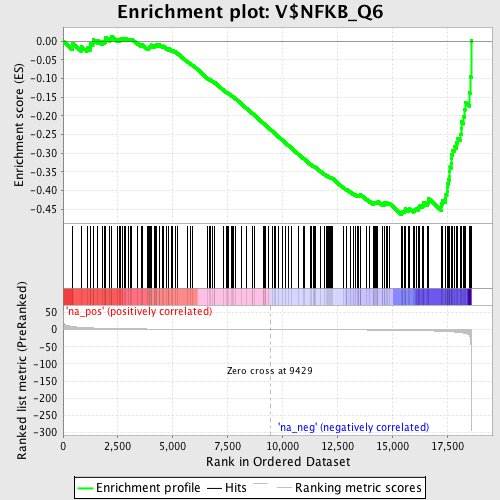
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMpreB_versus_WTpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFKB_Q6 |
| Enrichment Score (ES) | -0.46431786 |
| Normalized Enrichment Score (NES) | -1.5281821 |
| Nominal p-value | 0.021148037 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | DAP3 | 421 | 9.000 | -0.0051 | No | ||
| 2 | NLGN2 | 826 | 6.512 | -0.0142 | No | ||
| 3 | PCBP4 | 1090 | 5.475 | -0.0177 | No | ||
| 4 | RXRB | 1229 | 5.029 | -0.0153 | No | ||
| 5 | CD86 | 1255 | 4.945 | -0.0070 | No | ||
| 6 | TCEA2 | 1363 | 4.646 | -0.0036 | No | ||
| 7 | NUP62 | 1384 | 4.574 | 0.0043 | No | ||
| 8 | STIP1 | 1561 | 4.141 | 0.0029 | No | ||
| 9 | ELOVL6 | 1788 | 3.696 | -0.0021 | No | ||
| 10 | CHD4 | 1881 | 3.494 | -0.0003 | No | ||
| 11 | PABPC1 | 1934 | 3.403 | 0.0036 | No | ||
| 12 | HSD3B7 | 1943 | 3.391 | 0.0098 | No | ||
| 13 | GATA4 | 2122 | 3.119 | 0.0063 | No | ||
| 14 | XPO6 | 2186 | 3.012 | 0.0088 | No | ||
| 15 | RGL1 | 2210 | 2.986 | 0.0135 | No | ||
| 16 | PPP1R13B | 2463 | 2.657 | 0.0050 | No | ||
| 17 | PPARBP | 2585 | 2.535 | 0.0034 | No | ||
| 18 | HCFC1 | 2632 | 2.492 | 0.0058 | No | ||
| 19 | PSMA1 | 2688 | 2.448 | 0.0077 | No | ||
| 20 | SLC39A7 | 2803 | 2.343 | 0.0061 | No | ||
| 21 | NFKB2 | 2856 | 2.303 | 0.0078 | No | ||
| 22 | GPR150 | 2983 | 2.193 | 0.0053 | No | ||
| 23 | SIN3A | 3059 | 2.132 | 0.0054 | No | ||
| 24 | TOMM20 | 3132 | 2.079 | 0.0056 | No | ||
| 25 | FANCC | 3405 | 1.873 | -0.0055 | No | ||
| 26 | PLXDC2 | 3582 | 1.770 | -0.0116 | No | ||
| 27 | AMOTL1 | 3598 | 1.763 | -0.0089 | No | ||
| 28 | TNFSF15 | 3834 | 1.637 | -0.0184 | No | ||
| 29 | IER5 | 3899 | 1.602 | -0.0187 | No | ||
| 30 | NDRG2 | 3914 | 1.596 | -0.0164 | No | ||
| 31 | RFX5 | 3954 | 1.579 | -0.0154 | No | ||
| 32 | PTHLH | 4002 | 1.552 | -0.0149 | No | ||
| 33 | CXCL9 | 4028 | 1.545 | -0.0132 | No | ||
| 34 | CYP26A1 | 4030 | 1.544 | -0.0102 | No | ||
| 35 | NOS1 | 4151 | 1.498 | -0.0138 | No | ||
| 36 | HOXB3 | 4180 | 1.482 | -0.0124 | No | ||
| 37 | FGF17 | 4228 | 1.461 | -0.0121 | No | ||
| 38 | STC2 | 4247 | 1.452 | -0.0102 | No | ||
| 39 | TRIM47 | 4278 | 1.437 | -0.0090 | No | ||
| 40 | DDR1 | 4371 | 1.402 | -0.0112 | No | ||
| 41 | SP6 | 4373 | 1.401 | -0.0085 | No | ||
| 42 | SLAMF8 | 4538 | 1.338 | -0.0148 | No | ||
| 43 | SLC16A4 | 4554 | 1.333 | -0.0130 | No | ||
| 44 | BNC2 | 4711 | 1.271 | -0.0189 | No | ||
| 45 | IL1RN | 4811 | 1.229 | -0.0219 | No | ||
| 46 | CDK6 | 4816 | 1.226 | -0.0197 | No | ||
| 47 | BDNF | 4945 | 1.182 | -0.0243 | No | ||
| 48 | SUFU | 4998 | 1.168 | -0.0248 | No | ||
| 49 | CHX10 | 5103 | 1.139 | -0.0282 | No | ||
| 50 | TIAL1 | 5227 | 1.100 | -0.0327 | No | ||
| 51 | EHF | 5656 | 0.967 | -0.0541 | No | ||
| 52 | TNFSF18 | 5798 | 0.926 | -0.0599 | No | ||
| 53 | ADAMTS5 | 5908 | 0.895 | -0.0640 | No | ||
| 54 | VCAM1 | 6600 | 0.710 | -0.1001 | No | ||
| 55 | CYR61 | 6688 | 0.687 | -0.1035 | No | ||
| 56 | PCDH10 | 6714 | 0.682 | -0.1035 | No | ||
| 57 | OGG1 | 6817 | 0.655 | -0.1077 | No | ||
| 58 | TOP1 | 6905 | 0.631 | -0.1112 | No | ||
| 59 | IFNB1 | 6917 | 0.628 | -0.1106 | No | ||
| 60 | FKHL18 | 7298 | 0.537 | -0.1301 | No | ||
| 61 | CCL5 | 7441 | 0.500 | -0.1369 | No | ||
| 62 | MAG | 7494 | 0.485 | -0.1387 | No | ||
| 63 | NFAT5 | 7549 | 0.473 | -0.1407 | No | ||
| 64 | UBE2I | 7676 | 0.438 | -0.1467 | No | ||
| 65 | GABRE | 7718 | 0.427 | -0.1481 | No | ||
| 66 | CPLX2 | 7774 | 0.412 | -0.1503 | No | ||
| 67 | SOX10 | 7874 | 0.387 | -0.1549 | No | ||
| 68 | CLCN1 | 8128 | 0.326 | -0.1679 | No | ||
| 69 | TMEM27 | 8365 | 0.264 | -0.1802 | No | ||
| 70 | TCF1 | 8615 | 0.206 | -0.1933 | No | ||
| 71 | PTGIR | 8645 | 0.197 | -0.1945 | No | ||
| 72 | XPO1 | 8722 | 0.178 | -0.1983 | No | ||
| 73 | TAL1 | 9113 | 0.082 | -0.2193 | No | ||
| 74 | MAP3K8 | 9126 | 0.078 | -0.2198 | No | ||
| 75 | RIN2 | 9161 | 0.066 | -0.2215 | No | ||
| 76 | BFSP1 | 9231 | 0.050 | -0.2251 | No | ||
| 77 | HOXB9 | 9234 | 0.050 | -0.2251 | No | ||
| 78 | MNT | 9368 | 0.015 | -0.2323 | No | ||
| 79 | FOXD2 | 9549 | -0.035 | -0.2420 | No | ||
| 80 | CTGF | 9648 | -0.059 | -0.2472 | No | ||
| 81 | SLC6A12 | 9677 | -0.066 | -0.2486 | No | ||
| 82 | HOXB2 | 9801 | -0.100 | -0.2551 | No | ||
| 83 | TNFSF4 | 10014 | -0.157 | -0.2663 | No | ||
| 84 | PTGES | 10139 | -0.186 | -0.2726 | No | ||
| 85 | TCF15 | 10291 | -0.218 | -0.2804 | No | ||
| 86 | JAK3 | 10399 | -0.239 | -0.2857 | No | ||
| 87 | MOBKL2C | 10719 | -0.312 | -0.3024 | No | ||
| 88 | MLLT6 | 10950 | -0.374 | -0.3142 | No | ||
| 89 | CYBB | 10990 | -0.382 | -0.3155 | No | ||
| 90 | SEC14L2 | 11264 | -0.450 | -0.3295 | No | ||
| 91 | PDE3B | 11336 | -0.469 | -0.3324 | No | ||
| 92 | PTGS2 | 11414 | -0.487 | -0.3356 | No | ||
| 93 | CXCL2 | 11456 | -0.498 | -0.3368 | No | ||
| 94 | CGN | 11457 | -0.498 | -0.3359 | No | ||
| 95 | NFKBIB | 11524 | -0.516 | -0.3384 | No | ||
| 96 | WRN | 11741 | -0.574 | -0.3490 | No | ||
| 97 | TSLP | 11913 | -0.622 | -0.3571 | No | ||
| 98 | PURG | 12024 | -0.654 | -0.3618 | No | ||
| 99 | ARHGEF2 | 12030 | -0.656 | -0.3607 | No | ||
| 100 | FGF1 | 12092 | -0.674 | -0.3627 | No | ||
| 101 | BCL3 | 12157 | -0.691 | -0.3648 | No | ||
| 102 | UACA | 12208 | -0.705 | -0.3662 | No | ||
| 103 | PFN1 | 12242 | -0.713 | -0.3665 | No | ||
| 104 | PLA1A | 12276 | -0.725 | -0.3669 | No | ||
| 105 | C1QL1 | 12769 | -0.880 | -0.3919 | No | ||
| 106 | BCKDK | 12908 | -0.918 | -0.3975 | No | ||
| 107 | ASH1L | 12932 | -0.924 | -0.3970 | No | ||
| 108 | CSF2 | 13096 | -0.975 | -0.4039 | No | ||
| 109 | ATF5 | 13212 | -1.021 | -0.4081 | No | ||
| 110 | GRIK2 | 13305 | -1.054 | -0.4110 | No | ||
| 111 | HOXA11 | 13400 | -1.088 | -0.4140 | No | ||
| 112 | KLK9 | 13440 | -1.105 | -0.4140 | No | ||
| 113 | CACNA1E | 13481 | -1.118 | -0.4139 | No | ||
| 114 | TANK | 13533 | -1.141 | -0.4144 | No | ||
| 115 | GREM1 | 13540 | -1.145 | -0.4125 | No | ||
| 116 | IL23A | 13558 | -1.152 | -0.4112 | No | ||
| 117 | GNAO1 | 13825 | -1.256 | -0.4231 | No | ||
| 118 | CXCL10 | 13979 | -1.314 | -0.4288 | No | ||
| 119 | RND1 | 14154 | -1.381 | -0.4356 | No | ||
| 120 | PAPPA | 14160 | -1.382 | -0.4331 | No | ||
| 121 | RAI1 | 14203 | -1.401 | -0.4326 | No | ||
| 122 | CYLD | 14228 | -1.408 | -0.4312 | No | ||
| 123 | SEMA3B | 14303 | -1.438 | -0.4324 | No | ||
| 124 | FXYD2 | 14333 | -1.452 | -0.4311 | No | ||
| 125 | BCOR | 14348 | -1.458 | -0.4290 | No | ||
| 126 | GJB1 | 14552 | -1.546 | -0.4369 | No | ||
| 127 | GRK5 | 14565 | -1.551 | -0.4346 | No | ||
| 128 | PRKCD | 14653 | -1.601 | -0.4361 | No | ||
| 129 | SYT7 | 14655 | -1.603 | -0.4330 | No | ||
| 130 | NLK | 14717 | -1.635 | -0.4331 | No | ||
| 131 | TNIP1 | 14783 | -1.671 | -0.4334 | No | ||
| 132 | LRP1 | 14855 | -1.717 | -0.4338 | No | ||
| 133 | RGS3 | 15418 | -2.102 | -0.4602 | Yes | ||
| 134 | PCSK2 | 15448 | -2.126 | -0.4576 | Yes | ||
| 135 | ERN1 | 15489 | -2.159 | -0.4555 | Yes | ||
| 136 | RYBP | 15555 | -2.208 | -0.4547 | Yes | ||
| 137 | ASCL3 | 15591 | -2.239 | -0.4522 | Yes | ||
| 138 | GBA2 | 15614 | -2.266 | -0.4489 | Yes | ||
| 139 | MSC | 15745 | -2.375 | -0.4513 | Yes | ||
| 140 | AP1S2 | 15797 | -2.432 | -0.4493 | Yes | ||
| 141 | ICAM1 | 15958 | -2.602 | -0.4528 | Yes | ||
| 142 | CDC14A | 16035 | -2.699 | -0.4517 | Yes | ||
| 143 | ILK | 16089 | -2.770 | -0.4491 | Yes | ||
| 144 | NSD1 | 16180 | -2.884 | -0.4483 | Yes | ||
| 145 | PDE4C | 16213 | -2.922 | -0.4443 | Yes | ||
| 146 | REL | 16264 | -2.984 | -0.4411 | Yes | ||
| 147 | ATP1B3 | 16365 | -3.157 | -0.4404 | Yes | ||
| 148 | NR2F2 | 16413 | -3.231 | -0.4366 | Yes | ||
| 149 | ZADH2 | 16436 | -3.272 | -0.4313 | Yes | ||
| 150 | SLC11A2 | 16597 | -3.553 | -0.4330 | Yes | ||
| 151 | PLAU | 16648 | -3.652 | -0.4285 | Yes | ||
| 152 | HOXA7 | 16657 | -3.671 | -0.4218 | Yes | ||
| 153 | TRIB2 | 17245 | -5.062 | -0.4436 | Yes | ||
| 154 | P2RY10 | 17253 | -5.107 | -0.4340 | Yes | ||
| 155 | PTPRJ | 17302 | -5.236 | -0.4263 | Yes | ||
| 156 | DNAJB5 | 17432 | -5.603 | -0.4223 | Yes | ||
| 157 | STAT6 | 17447 | -5.689 | -0.4118 | Yes | ||
| 158 | WNT10A | 17497 | -5.896 | -0.4029 | Yes | ||
| 159 | ZHX2 | 17508 | -5.940 | -0.3918 | Yes | ||
| 160 | LTB | 17536 | -6.038 | -0.3814 | Yes | ||
| 161 | ACTR1A | 17573 | -6.128 | -0.3713 | Yes | ||
| 162 | RAP2C | 17622 | -6.298 | -0.3615 | Yes | ||
| 163 | NFKBIA | 17625 | -6.306 | -0.3492 | Yes | ||
| 164 | IL2RA | 17631 | -6.321 | -0.3370 | Yes | ||
| 165 | PAFAH1B3 | 17686 | -6.463 | -0.3273 | Yes | ||
| 166 | ARPC5 | 17697 | -6.518 | -0.3150 | Yes | ||
| 167 | TGFB1 | 17710 | -6.583 | -0.3027 | Yes | ||
| 168 | LASP1 | 17764 | -6.772 | -0.2923 | Yes | ||
| 169 | CD83 | 17842 | -7.118 | -0.2824 | Yes | ||
| 170 | BLR1 | 17922 | -7.579 | -0.2718 | Yes | ||
| 171 | RUNX1 | 17993 | -7.864 | -0.2602 | Yes | ||
| 172 | SATB1 | 18110 | -8.715 | -0.2493 | Yes | ||
| 173 | BMF | 18144 | -9.009 | -0.2334 | Yes | ||
| 174 | LMO4 | 18154 | -9.070 | -0.2160 | Yes | ||
| 175 | ATP1B1 | 18264 | -10.083 | -0.2021 | Yes | ||
| 176 | EPHA2 | 18292 | -10.437 | -0.1831 | Yes | ||
| 177 | SAMSN1 | 18320 | -10.839 | -0.1632 | Yes | ||
| 178 | SOX5 | 18531 | -18.503 | -0.1382 | Yes | ||
| 179 | IL4I1 | 18552 | -22.957 | -0.0942 | Yes | ||
| 180 | BHLHB2 | 18597 | -49.644 | 0.0010 | Yes |
