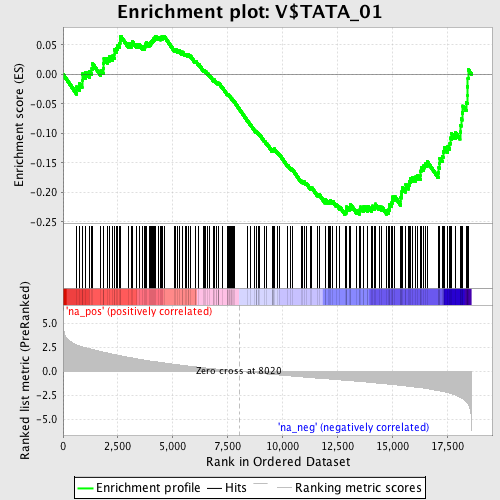
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set03_wtNotch_versus_truncNotch |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$TATA_01 |
| Enrichment Score (ES) | -0.23746556 |
| Normalized Enrichment Score (NES) | -1.2325252 |
| Nominal p-value | 0.06680585 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ANKMY2 | 623 | 2.757 | -0.0213 | No | ||
| 2 | SFRS3 | 745 | 2.665 | -0.0158 | No | ||
| 3 | H3F3B | 864 | 2.572 | -0.0105 | No | ||
| 4 | NASP | 872 | 2.569 | 0.0008 | No | ||
| 5 | EIF4A2 | 1023 | 2.470 | 0.0039 | No | ||
| 6 | TTR | 1210 | 2.350 | 0.0044 | No | ||
| 7 | PPP1R10 | 1312 | 2.293 | 0.0094 | No | ||
| 8 | ITPR1 | 1346 | 2.271 | 0.0179 | No | ||
| 9 | COL13A1 | 1724 | 2.063 | 0.0068 | No | ||
| 10 | CFL2 | 1833 | 2.009 | 0.0100 | No | ||
| 11 | MYH4 | 1846 | 2.002 | 0.0185 | No | ||
| 12 | MAT2A | 1861 | 1.992 | 0.0268 | No | ||
| 13 | HSPB2 | 2013 | 1.912 | 0.0273 | No | ||
| 14 | BMI1 | 2106 | 1.860 | 0.0307 | No | ||
| 15 | CRYAB | 2249 | 1.793 | 0.0311 | No | ||
| 16 | HOXD9 | 2337 | 1.749 | 0.0344 | No | ||
| 17 | SIM1 | 2340 | 1.747 | 0.0422 | No | ||
| 18 | MID1 | 2443 | 1.701 | 0.0444 | No | ||
| 19 | DDX3X | 2492 | 1.681 | 0.0494 | No | ||
| 20 | H2AFZ | 2566 | 1.649 | 0.0529 | No | ||
| 21 | DDX5 | 2595 | 1.638 | 0.0588 | No | ||
| 22 | EEF1A1 | 2625 | 1.619 | 0.0646 | No | ||
| 23 | TMPO | 2980 | 1.460 | 0.0520 | No | ||
| 24 | GRIN2B | 3102 | 1.411 | 0.0519 | No | ||
| 25 | RHOBTB1 | 3141 | 1.396 | 0.0561 | No | ||
| 26 | NEFH | 3350 | 1.313 | 0.0508 | No | ||
| 27 | KIRREL2 | 3460 | 1.268 | 0.0506 | No | ||
| 28 | FOXP3 | 3618 | 1.207 | 0.0476 | No | ||
| 29 | AMPD1 | 3731 | 1.163 | 0.0468 | No | ||
| 30 | NEUROG2 | 3744 | 1.159 | 0.0514 | No | ||
| 31 | NANOS1 | 3795 | 1.141 | 0.0539 | No | ||
| 32 | GTF2A1 | 3918 | 1.099 | 0.0523 | No | ||
| 33 | RUNX2 | 3986 | 1.071 | 0.0535 | No | ||
| 34 | SLC25A4 | 4028 | 1.055 | 0.0560 | No | ||
| 35 | PTP4A1 | 4082 | 1.036 | 0.0579 | No | ||
| 36 | EGFL7 | 4128 | 1.022 | 0.0601 | No | ||
| 37 | RASGRP3 | 4184 | 1.005 | 0.0616 | No | ||
| 38 | JARID2 | 4213 | 0.998 | 0.0647 | No | ||
| 39 | GPRC5D | 4335 | 0.959 | 0.0624 | No | ||
| 40 | RLN3 | 4418 | 0.932 | 0.0622 | No | ||
| 41 | CXCR4 | 4476 | 0.915 | 0.0633 | No | ||
| 42 | STC1 | 4549 | 0.889 | 0.0634 | No | ||
| 43 | PURA | 4614 | 0.866 | 0.0639 | No | ||
| 44 | GSC | 5084 | 0.727 | 0.0417 | No | ||
| 45 | PRDX1 | 5143 | 0.709 | 0.0418 | No | ||
| 46 | MITF | 5235 | 0.686 | 0.0400 | No | ||
| 47 | PLAG1 | 5286 | 0.670 | 0.0403 | No | ||
| 48 | RHOA | 5418 | 0.627 | 0.0360 | No | ||
| 49 | FGF4 | 5438 | 0.623 | 0.0378 | No | ||
| 50 | HIST1H2AA | 5560 | 0.592 | 0.0339 | No | ||
| 51 | GRID2 | 5610 | 0.576 | 0.0339 | No | ||
| 52 | IL11 | 5694 | 0.551 | 0.0319 | No | ||
| 53 | DIO2 | 5703 | 0.549 | 0.0340 | No | ||
| 54 | CA2 | 5798 | 0.525 | 0.0312 | No | ||
| 55 | EDN1 | 6015 | 0.473 | 0.0217 | No | ||
| 56 | MYF6 | 6049 | 0.464 | 0.0220 | No | ||
| 57 | STAC | 6176 | 0.429 | 0.0171 | No | ||
| 58 | HIST1H1C | 6411 | 0.371 | 0.0061 | No | ||
| 59 | SCN8A | 6433 | 0.367 | 0.0066 | No | ||
| 60 | FGF20 | 6504 | 0.348 | 0.0044 | No | ||
| 61 | NR0B2 | 6589 | 0.331 | 0.0013 | No | ||
| 62 | NOG | 6679 | 0.308 | -0.0021 | No | ||
| 63 | CDX1 | 6862 | 0.264 | -0.0108 | No | ||
| 64 | ASB16 | 6870 | 0.263 | -0.0100 | No | ||
| 65 | PANK1 | 6882 | 0.261 | -0.0094 | No | ||
| 66 | MMP20 | 6991 | 0.234 | -0.0142 | No | ||
| 67 | SLC2A4 | 6993 | 0.234 | -0.0132 | No | ||
| 68 | BHLHB5 | 7063 | 0.218 | -0.0159 | No | ||
| 69 | OLIG3 | 7086 | 0.215 | -0.0162 | No | ||
| 70 | EGR2 | 7090 | 0.214 | -0.0153 | No | ||
| 71 | CRABP2 | 7096 | 0.213 | -0.0147 | No | ||
| 72 | TSC1 | 7262 | 0.172 | -0.0228 | No | ||
| 73 | IL22 | 7486 | 0.120 | -0.0344 | No | ||
| 74 | SEPP1 | 7503 | 0.115 | -0.0347 | No | ||
| 75 | WSB2 | 7509 | 0.115 | -0.0345 | No | ||
| 76 | ARHGEF2 | 7520 | 0.114 | -0.0345 | No | ||
| 77 | GADD45G | 7569 | 0.104 | -0.0366 | No | ||
| 78 | HOXD4 | 7586 | 0.100 | -0.0371 | No | ||
| 79 | CSH2 | 7645 | 0.088 | -0.0398 | No | ||
| 80 | BZW2 | 7691 | 0.078 | -0.0419 | No | ||
| 81 | MYH3 | 7695 | 0.078 | -0.0417 | No | ||
| 82 | DKK1 | 7726 | 0.069 | -0.0430 | No | ||
| 83 | GDF8 | 7769 | 0.061 | -0.0450 | No | ||
| 84 | CKB | 7800 | 0.055 | -0.0464 | No | ||
| 85 | PLUNC | 8398 | -0.082 | -0.0784 | No | ||
| 86 | TNFSF10 | 8537 | -0.113 | -0.0854 | No | ||
| 87 | ADAMTS1 | 8739 | -0.151 | -0.0956 | No | ||
| 88 | FGF16 | 8798 | -0.162 | -0.0980 | No | ||
| 89 | ESRRG | 8806 | -0.163 | -0.0977 | No | ||
| 90 | OLIG2 | 8807 | -0.163 | -0.0969 | No | ||
| 91 | DUOX2 | 8896 | -0.185 | -0.1009 | No | ||
| 92 | SCHIP1 | 8902 | -0.185 | -0.1003 | No | ||
| 93 | HOXC4 | 8957 | -0.197 | -0.1023 | No | ||
| 94 | ELAVL3 | 9184 | -0.248 | -0.1135 | No | ||
| 95 | SPRR1B | 9284 | -0.274 | -0.1176 | No | ||
| 96 | SLC34A1 | 9532 | -0.326 | -0.1295 | No | ||
| 97 | SOSTDC1 | 9542 | -0.328 | -0.1285 | No | ||
| 98 | TRDN | 9551 | -0.329 | -0.1275 | No | ||
| 99 | TMEPAI | 9583 | -0.337 | -0.1276 | No | ||
| 100 | ADAM11 | 9605 | -0.341 | -0.1272 | No | ||
| 101 | ACTA1 | 9616 | -0.343 | -0.1262 | No | ||
| 102 | CILP | 9759 | -0.372 | -0.1322 | No | ||
| 103 | GBE1 | 9871 | -0.390 | -0.1365 | No | ||
| 104 | CYP1A2 | 10240 | -0.461 | -0.1544 | No | ||
| 105 | RGS4 | 10367 | -0.483 | -0.1590 | No | ||
| 106 | NPTX1 | 10441 | -0.496 | -0.1607 | No | ||
| 107 | DKK2 | 10858 | -0.574 | -0.1807 | No | ||
| 108 | KRTAP6-2 | 10923 | -0.588 | -0.1815 | No | ||
| 109 | DES | 10986 | -0.600 | -0.1821 | No | ||
| 110 | CNN1 | 11078 | -0.619 | -0.1843 | No | ||
| 111 | LTA | 11275 | -0.657 | -0.1919 | No | ||
| 112 | BACH1 | 11341 | -0.667 | -0.1924 | No | ||
| 113 | MT4 | 11599 | -0.712 | -0.2031 | No | ||
| 114 | HOXA11 | 11663 | -0.721 | -0.2033 | No | ||
| 115 | RGS1 | 11939 | -0.769 | -0.2147 | No | ||
| 116 | RPS3 | 11949 | -0.770 | -0.2117 | No | ||
| 117 | AQP1 | 12084 | -0.797 | -0.2154 | No | ||
| 118 | S100A4 | 12142 | -0.805 | -0.2148 | No | ||
| 119 | FGFRL1 | 12186 | -0.813 | -0.2135 | No | ||
| 120 | ABCA1 | 12299 | -0.834 | -0.2157 | No | ||
| 121 | LHX6 | 12459 | -0.863 | -0.2205 | No | ||
| 122 | CRYBA1 | 12614 | -0.892 | -0.2248 | No | ||
| 123 | TTC17 | 12849 | -0.935 | -0.2332 | Yes | ||
| 124 | ATOH1 | 12896 | -0.944 | -0.2314 | Yes | ||
| 125 | KBTBD5 | 12917 | -0.947 | -0.2282 | Yes | ||
| 126 | SOX3 | 12930 | -0.950 | -0.2246 | Yes | ||
| 127 | MT3 | 13038 | -0.968 | -0.2260 | Yes | ||
| 128 | NDRG2 | 13101 | -0.980 | -0.2249 | Yes | ||
| 129 | DIRAS1 | 13102 | -0.980 | -0.2204 | Yes | ||
| 130 | HIST1H1T | 13386 | -1.036 | -0.2311 | Yes | ||
| 131 | TNNC2 | 13496 | -1.056 | -0.2322 | Yes | ||
| 132 | KRTAP21-1 | 13529 | -1.066 | -0.2291 | Yes | ||
| 133 | TCTA | 13537 | -1.067 | -0.2246 | Yes | ||
| 134 | CSH1 | 13696 | -1.093 | -0.2283 | Yes | ||
| 135 | HDLBP | 13707 | -1.096 | -0.2238 | Yes | ||
| 136 | SERPINH1 | 13877 | -1.127 | -0.2279 | Yes | ||
| 137 | BNIP3 | 13886 | -1.129 | -0.2232 | Yes | ||
| 138 | ECE2 | 14060 | -1.165 | -0.2273 | Yes | ||
| 139 | SUPT4H1 | 14080 | -1.168 | -0.2230 | Yes | ||
| 140 | TSHB | 14206 | -1.197 | -0.2244 | Yes | ||
| 141 | PABPC4 | 14220 | -1.200 | -0.2196 | Yes | ||
| 142 | CKM | 14402 | -1.244 | -0.2238 | Yes | ||
| 143 | CCL5 | 14526 | -1.272 | -0.2247 | Yes | ||
| 144 | HOXC5 | 14755 | -1.315 | -0.2311 | Yes | ||
| 145 | COL1A1 | 14840 | -1.333 | -0.2296 | Yes | ||
| 146 | TPM1 | 14873 | -1.341 | -0.2253 | Yes | ||
| 147 | MYCT1 | 14884 | -1.344 | -0.2197 | Yes | ||
| 148 | RBP2 | 14963 | -1.364 | -0.2177 | Yes | ||
| 149 | NR4A1 | 14996 | -1.372 | -0.2133 | Yes | ||
| 150 | ZFP36L2 | 15010 | -1.375 | -0.2077 | Yes | ||
| 151 | MYOZ2 | 15116 | -1.398 | -0.2071 | Yes | ||
| 152 | SOX5 | 15391 | -1.461 | -0.2153 | Yes | ||
| 153 | HOXA10 | 15394 | -1.462 | -0.2088 | Yes | ||
| 154 | UCN | 15412 | -1.467 | -0.2030 | Yes | ||
| 155 | CCL7 | 15440 | -1.474 | -0.1978 | Yes | ||
| 156 | TGM3 | 15449 | -1.477 | -0.1915 | Yes | ||
| 157 | PCDH8 | 15599 | -1.517 | -0.1927 | Yes | ||
| 158 | LIF | 15622 | -1.523 | -0.1870 | Yes | ||
| 159 | HNRPA0 | 15760 | -1.560 | -0.1874 | Yes | ||
| 160 | COL10A1 | 15776 | -1.567 | -0.1811 | Yes | ||
| 161 | CDC25B | 15829 | -1.580 | -0.1767 | Yes | ||
| 162 | SMYD1 | 15925 | -1.607 | -0.1746 | Yes | ||
| 163 | TCF7 | 16050 | -1.644 | -0.1739 | Yes | ||
| 164 | HIST1H2BA | 16142 | -1.667 | -0.1712 | Yes | ||
| 165 | CLCN1 | 16286 | -1.705 | -0.1712 | Yes | ||
| 166 | BRUNOL4 | 16292 | -1.707 | -0.1638 | Yes | ||
| 167 | NNT | 16315 | -1.716 | -0.1572 | Yes | ||
| 168 | FSHB | 16424 | -1.753 | -0.1551 | Yes | ||
| 169 | TNFRSF17 | 16494 | -1.778 | -0.1507 | Yes | ||
| 170 | CX3CL1 | 16590 | -1.816 | -0.1477 | Yes | ||
| 171 | ADAMTS15 | 17106 | -2.023 | -0.1664 | Yes | ||
| 172 | RPL10 | 17126 | -2.030 | -0.1582 | Yes | ||
| 173 | MMP10 | 17152 | -2.036 | -0.1503 | Yes | ||
| 174 | POU3F4 | 17169 | -2.043 | -0.1419 | Yes | ||
| 175 | EEF2 | 17287 | -2.099 | -0.1388 | Yes | ||
| 176 | GCH1 | 17322 | -2.113 | -0.1310 | Yes | ||
| 177 | ABLIM1 | 17364 | -2.137 | -0.1235 | Yes | ||
| 178 | MIP | 17535 | -2.237 | -0.1226 | Yes | ||
| 179 | ASB4 | 17628 | -2.289 | -0.1172 | Yes | ||
| 180 | NPPA | 17648 | -2.303 | -0.1078 | Yes | ||
| 181 | PDLIM7 | 17707 | -2.343 | -0.1003 | Yes | ||
| 182 | AGT | 17873 | -2.472 | -0.0980 | Yes | ||
| 183 | MRPS18B | 18090 | -2.693 | -0.0975 | Yes | ||
| 184 | HIST1H1B | 18117 | -2.734 | -0.0865 | Yes | ||
| 185 | PDYN | 18140 | -2.755 | -0.0752 | Yes | ||
| 186 | PPARG | 18183 | -2.809 | -0.0647 | Yes | ||
| 187 | MAP2K5 | 18221 | -2.871 | -0.0537 | Yes | ||
| 188 | CSNK1E | 18377 | -3.173 | -0.0477 | Yes | ||
| 189 | IL10 | 18421 | -3.274 | -0.0352 | Yes | ||
| 190 | HIST1H1A | 18428 | -3.292 | -0.0206 | Yes | ||
| 191 | SCAMP3 | 18450 | -3.350 | -0.0065 | Yes | ||
| 192 | KLF2 | 18472 | -3.402 | 0.0078 | Yes |
